HOXA5 Is Recognized as a Prognostic-Related Biomarker and Promotes Glioma Progression Through Affecting Cell Cycle
- PMID: 34485110
- PMCID: PMC8416157
- DOI: 10.3389/fonc.2021.633430
HOXA5 Is Recognized as a Prognostic-Related Biomarker and Promotes Glioma Progression Through Affecting Cell Cycle
Abstract
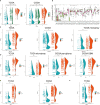
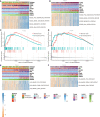
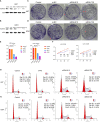
Glioma is malignant tumor derives from glial cells in the central nervous system. High-grade glioma shows aggressive growth pattern, and conventional treatments, such as surgical removal and chemo-radiotherapy, archive limitation in the interference of this process. In this work, HOXA5, from the HOX family, was identified as a glioma cell proliferation-associated factor by investigating its feature in the TCGA and CGGA data set. High HOXA5 expression samples contain unfavorable clinical features of glioma, including IDH wild type, un-methylated MGMT status, non-codeletion 1p19q status, malignant molecular subtype. Survival analysis indicates that high HOXA5 expression samples are associated with worse clinical outcome. The CNVs and SNPs profile difference further confirmed the enrichment of glioma aggressive related biomarkers. In the meantime, the activation of DNA damage repair-related pathways and TP53-related pathways is also related to HOXA5 expression. In cell lines, U87MG and U251, by interfering HOXA5 expression significantly inhibit glioma progression and apoptosis, and cell cycle is arrested at the G2/M phase. Collectively, increased HOXA5 expression can promote glioma progression via affecting glioma cell proliferation.
Keywords: cell cycle; cell proliferation; glioma; homeobox A5; prognosis.
Copyright © 2021 Ding, Chen, Bie, Piao and Cheng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
Abnormal expression of HOXD11 promotes the malignant behavior of glioma cells and leads to poor prognosis of glioma patients.PeerJ. 2021 Feb 8;9:e10820. doi: 10.7717/peerj.10820. eCollection 2021. PeerJ. 2021. PMID: 33614284 Free PMC article.
-
A nuclear transport-related gene signature combined with IDH mutation and 1p/19q codeletion better predicts the prognosis of glioma patients.BMC Cancer. 2020 Nov 9;20(1):1072. doi: 10.1186/s12885-020-07552-3. BMC Cancer. 2020. PMID: 33167941 Free PMC article.
-
HOXA5 indicates poor prognosis and suppresses cell proliferation by regulating p21 expression in non small cell lung cancer.Tumour Biol. 2015 May;36(5):3521-31. doi: 10.1007/s13277-014-2988-4. Epub 2014 Dec 31. Tumour Biol. 2015. PMID: 25549794
-
ESPL1 Is a Novel Prognostic Biomarker Associated With the Malignant Features of Glioma.Front Genet. 2021 Aug 26;12:666106. doi: 10.3389/fgene.2021.666106. eCollection 2021. Front Genet. 2021. PMID: 34512713 Free PMC article.
-
Increased HOXA5 expression provides a selective advantage for gain of whole chromosome 7 in IDH wild-type glioblastoma.Genes Dev. 2018 Apr 1;32(7-8):512-523. doi: 10.1101/gad.312157.118. Epub 2018 Apr 9. Genes Dev. 2018. PMID: 29632085 Free PMC article.
Cited by
-
Identification of Gender- and Subtype-Specific Gene Expression Associated with Patient Survival in Low-Grade and Anaplastic Glioma in Connection with Steroid Signaling.Cancers (Basel). 2022 Aug 25;14(17):4114. doi: 10.3390/cancers14174114. Cancers (Basel). 2022. PMID: 36077653 Free PMC article.
-
Super-enhancer-driven SLCO4A1-AS1 is a new biomarker and a promising therapeutic target in glioblastoma.Sci Rep. 2025 Jan 6;15(1):954. doi: 10.1038/s41598-024-82109-z. Sci Rep. 2025. PMID: 39762261 Free PMC article.
-
The Transcription Factor HOXA5: Novel Insights into Metabolic Diseases and Adipose Tissue Dysfunction.Cells. 2023 Aug 18;12(16):2090. doi: 10.3390/cells12162090. Cells. 2023. PMID: 37626900 Free PMC article. Review.
-
HOXA10 and HOXA11 in Human Endometrial Benign Disorders: Unraveling Molecular Pathways and Their Impact on Reproduction.Biomolecules. 2025 Apr 10;15(4):563. doi: 10.3390/biom15040563. Biomolecules. 2025. PMID: 40305321 Free PMC article. Review.
-
Protein Quality Control in Glioblastoma: A Review of the Current Literature with New Perspectives on Therapeutic Targets.Int J Mol Sci. 2022 Aug 27;23(17):9734. doi: 10.3390/ijms23179734. Int J Mol Sci. 2022. PMID: 36077131 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

