Podoplanin drives dedifferentiation and amoeboid invasion of melanoma
- PMID: 34485858
- PMCID: PMC8405990
- DOI: 10.1016/j.isci.2021.102976
Podoplanin drives dedifferentiation and amoeboid invasion of melanoma
Abstract
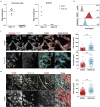
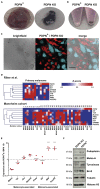
Melanoma is an aggressive skin cancer developing from melanocytes, frequently resulting in metastatic disease. Melanoma cells utilize amoeboid migration as mode of local invasion. Amoeboid invasion is characterized by rounded cell morphology and high actomyosin contractility driven by Rho GTPase signalling. Migrastatic drugs targeting actin polymerization and contractility are therefore a promising treatment option for metastatic melanoma. To predict amoeboid invasion and metastatic potential, biomarkers functionally linked to contractility pathways are needed. The glycoprotein podoplanin drives actomyosin contractility in lymphoid fibroblasts and is overexpressed in many cancers. We show that podoplanin enhances amoeboid invasion in melanoma. Podoplanin expression in murine melanoma drives rounded cell morphology, increasing motility, and invasion in vivo. Podoplanin expression is increased in a subset of dedifferentiated human melanoma, and in vitro is sufficient to upregulate melanoma-associated marker Pou3f2/Brn2. Together, our data define podoplanin as a functional biomarker for dedifferentiated invasive melanoma and a promising migrastatic therapeutic target.
Keywords: Cell biology; Molecular biology; Transcriptomics.
© 2021 The Author(s).
Figures




References
-
- Cantelli G., Orgaz J.L., Rodriguez-Hernandez I., Karagiannis P., Maiques O., Matias-Guiu X., Nestle F.O., Marti R.M., Karagiannis S.N., Sanz-Moreno V. TGF-β-Induced transcription sustains amoeboid melanoma migration and dissemination. Curr. Biol. 2015;25:2899–2914. doi: 10.1016/j.cub.2015.09.054. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

