Information content differentiates enhancers from silencers in mouse photoreceptors
- PMID: 34486522
- PMCID: PMC8492058
- DOI: 10.7554/eLife.67403
Information content differentiates enhancers from silencers in mouse photoreceptors
Abstract
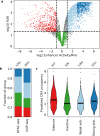
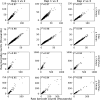
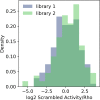
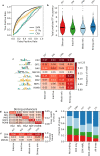
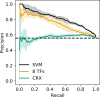
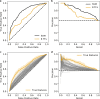
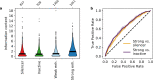
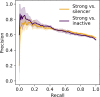
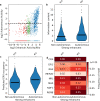
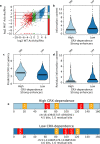
Enhancers and silencers often depend on the same transcription factors (TFs) and are conflated in genomic assays of TF binding or chromatin state. To identify sequence features that distinguish enhancers and silencers, we assayed massively parallel reporter libraries of genomic sequences targeted by the photoreceptor TF cone-rod homeobox (CRX) in mouse retinas. Both enhancers and silencers contain more TF motifs than inactive sequences, but relative to silencers, enhancers contain motifs from a more diverse collection of TFs. We developed a measure of information content that describes the number and diversity of motifs in a sequence and found that, while both enhancers and silencers depend on CRX motifs, enhancers have higher information content. The ability of information content to distinguish enhancers and silencers targeted by the same TF illustrates how motif context determines the activity of cis-regulatory sequences.
Keywords: computational biology; enhancers; genetics; genomics; information theory; massively parallel reporter assays; mouse; silencers; systems biology.
Plain language summary
Different cell types are established by activating and repressing the activity of specific sets of genes, a process controlled by proteins called transcription factors. Transcription factors work by recognizing and binding short stretches of DNA in parts of the genome called cis-regulatory sequences. A cis-regulatory sequence that increases the activity of a gene when bound by transcription factors is called an enhancer, while a sequence that causes a decrease in gene activity is called a silencer. To establish a cell type, a particular transcription factor will act on both enhancers and silencers that control the activity of different genes. For example, the transcription factor cone-rod homeobox (CRX) is critical for specifying different types of cells in the retina, and it acts on both enhancers and silencers. In rod photoreceptors, CRX activates rod genes by binding their enhancers, while repressing cone photoreceptor genes by binding their silencers. However, CRX always recognizes and binds to the same DNA sequence, known as its binding site, making it unclear why some cis-regulatory sequences bound to CRX act as silencers, while others act as enhancers. Friedman et al. sought to understand how enhancers and silencers, both bound by CRX, can have different effects on the genes they control. Since both enhancers and silencers contain CRX binding sites, the difference between the two must lie in the sequence of the DNA surrounding these binding sites. Using retinas that have been explanted from mice and kept alive in the laboratory, Friedman et al. tested the activity of thousands of CRX-binding sequences from the mouse genome. This showed that both enhancers and silencers have more copies of CRX-binding sites than sequences of the genome that are inactive. Additionally, the results revealed that enhancers have a diverse collection of binding sites for other transcription factors, while silencers do not. Friedman et al. developed a new metric they called information content, which captures the diverse combinations of different transcription binding sites that cis-regulatory sequences can have. Using this metric, Friedman et al. showed that it is possible to distinguish enhancers from silencers based on their information content. It is critical to understand how the DNA sequences of cis-regulatory regions determine their activity, because mutations in these regions of the genome can cause disease. However, since every person has thousands of benign mutations in cis-regulatory sequences, it is a challenge to identify specific disease-causing mutations, which are relatively rare. One long-term goal of models of enhancers and silencers, such as Friedman et al.’s information content model, is to understand how mutations can affect cis-regulatory sequences, and, in some cases, lead to disease.
© 2021, Friedman et al.
Conflict of interest statement
RF, DG, CM, JC, BC, MW No competing interests declared
Figures

References
-
- Andzelm MM, Cherry TJ, Harmin DA, Boeke AC, Lee C, Hemberg M, Pawlyk B, Malik AN, Flavell SW, Sandberg MA, Raviola E, Greenberg ME. MEF2D drives photoreceptor development through a genome-wide competition for tissue-specific enhancers. Neuron. 2015;86:247–263. doi: 10.1016/j.neuron.2015.02.038. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

