The fall and rise of group B Streptococcus in dairy cattle: reintroduction due to human-to-cattle host jumps?
- PMID: 34486971
- PMCID: PMC8715428
- DOI: 10.1099/mgen.0.000648
The fall and rise of group B Streptococcus in dairy cattle: reintroduction due to human-to-cattle host jumps?
Abstract
Group B
Keywords: Streptococcus agalactiae; anthroponosis; emergence; host adaptation; plasmid; reverse zoonosis.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures
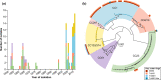
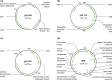
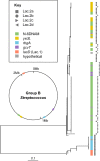

References
-
- Barkham T, Zadoks RN, Azmai MNA, Baker S, Bich VTN, et al. One hypervirulent clone, sequence type 283, accounts for a large proportion of invasive Streptococcus agalactiae isolated from humans and diseased tilapia in Southeast Asia. PLoS Negl Trop Dis. 2019;13:e0007421. doi: 10.1371/journal.pntd.0007421. - DOI - PMC - PubMed
-
- van der Mee-Marquet N, Fourny L, Arnault L, Domelier AS, Salloum M, et al. Molecular characterization of human-colonizing Streptococcus agalactiae strains isolated from throat, skin, anal margin, and genital body sites. J Clin Microbiol. 2008;46:2906–2911. doi: 10.1128/JCM.00421-08. - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

