GDF15 induces immunosuppression via CD48 on regulatory T cells in hepatocellular carcinoma
- PMID: 34489334
- PMCID: PMC8422483
- DOI: 10.1136/jitc-2021-002787
GDF15 induces immunosuppression via CD48 on regulatory T cells in hepatocellular carcinoma
Abstract
Background: A better understanding of the molecular mechanisms that manifest in the immunosuppressive tumor microenvironment (TME) is crucial for developing more efficacious immunotherapies for hepatocellular carcinoma (HCC), which has a poor response to current immunotherapies. Regulatory T (Treg) cells are key mediators of HCC-associated immunosuppression. We investigated the selective mechanism exploited by HCC that lead to Treg cells expansion and to find more efficacious immunotherapies.
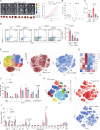
Methods: We used matched tumor tissues and blood samples from 150 patients with HCC to identify key factors of Treg cells expansion. We used mass cytometry (CyTOF) and orthotopic cancer mouse models to analyze overall immunological changes after growth differentiation factor 15 (GDF15) gene ablation in HCC. We used flow cytometry, coimmunoprecipitation, RNA sequencing, mass spectrum, chromatin immunoprecipitation and Gdf15-/-, OT-I and GFP transgenic mice to demonstrate the effects of GDF15 on Treg cells and related molecular mechanism. We used hybridoma technology to generate monoclonal antibody to block GDF15 and evaluate its effects on HCC-associated immunosuppression.
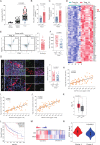
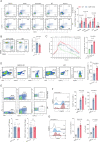
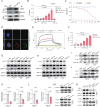
Results: GDF15 is positively associated with the elevation of Treg cell frequencies in patients wih HCC. Gene ablation of GDF15 in HCC can convert an immunosuppressive TME to an inflammatory state. GDF15 promotes the generation of peripherally derived inducible Treg (iTreg) cells and enhances the suppressive function of natural Treg (nTreg) cells by interacting with a previously unrecognized receptor CD48 on T cells and thus downregulates STUB1, an E3 ligase that mediates forkhead box P3 (FOXP3) protein degradation. GDF15 neutralizing antibody effectively eradicates HCC and augments the antitumor immunity in mouse.
Conclusions: Our results reveal the generation and function enhancement of Treg cells induced by GDF15 is a new mechanism for HCC-related immunosuppression. CD48 is the first discovered receptor of GDF15 in the immune system which provide the possibility to solve the molecular mechanism of the immunomodulatory function of GDF15. The therapeutic GDF15 blockade achieves HCC clearance without obvious adverse events.
Keywords: biomarkers; immune tolerance; immunotherapy; tumor; tumor escape; tumor microenvironment.
© Author(s) (or their employer(s)) 2021. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
