Nrf1 promotes heart regeneration and repair by regulating proteostasis and redox balance
- PMID: 34489413
- PMCID: PMC8421386
- DOI: 10.1038/s41467-021-25653-w
Nrf1 promotes heart regeneration and repair by regulating proteostasis and redox balance
Abstract
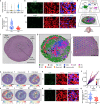
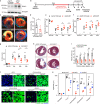
Following injury, cells in regenerative tissues have the ability to regrow. The mechanisms whereby regenerating cells adapt to injury-induced stress conditions and activate the regenerative program remain to be defined. Here, using the mammalian neonatal heart regeneration model, we show that Nrf1, a stress-responsive transcription factor encoded by the Nuclear Factor Erythroid 2 Like 1 (Nfe2l1) gene, is activated in regenerating cardiomyocytes. Genetic deletion of Nrf1 prevented regenerating cardiomyocytes from activating a transcriptional program required for heart regeneration. Conversely, Nrf1 overexpression protected the adult mouse heart from ischemia/reperfusion (I/R) injury. Nrf1 also protected human induced pluripotent stem cell-derived cardiomyocytes from doxorubicin-induced cardiotoxicity and other cardiotoxins. The protective function of Nrf1 is mediated by a dual stress response mechanism involving activation of the proteasome and redox balance. Our findings reveal that the adaptive stress response mechanism mediated by Nrf1 is required for neonatal heart regeneration and confers cardioprotection in the adult heart.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests as defined by Nature Research.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

