Brown adipose tissue monocytes support tissue expansion
- PMID: 34489438
- PMCID: PMC8421389
- DOI: 10.1038/s41467-021-25616-1
Brown adipose tissue monocytes support tissue expansion
Abstract
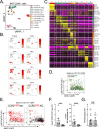
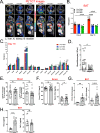
Monocytes are part of the mononuclear phagocytic system. Monocytes play a central role during inflammatory conditions and a better understanding of their dynamics might open therapeutic opportunities. In the present study, we focused on the characterization and impact of monocytes on brown adipose tissue (BAT) functions during tissue remodeling. Single-cell RNA sequencing analysis of BAT immune cells uncovered a large diversity in monocyte and macrophage populations. Fate-mapping experiments demonstrated that the BAT macrophage pool requires constant replenishment from monocytes. Using a genetic model of BAT expansion, we found that brown fat monocyte numbers were selectively increased in this scenario. This observation was confirmed using a CCR2-binding radiotracer and positron emission tomography. Importantly, in line with their tissue recruitment, blood monocyte counts were decreased while bone marrow hematopoiesis was not affected. Monocyte depletion prevented brown adipose tissue expansion and altered its architecture. Podoplanin engagement is strictly required for BAT expansion. Together, these data redefine the diversity of immune cells in the BAT and emphasize the role of monocyte recruitment for tissue remodeling.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Comment in
-
Monocytes involved in BAT expansion.Nat Rev Endocrinol. 2021 Dec;17(12):706. doi: 10.1038/s41574-021-00571-5. Nat Rev Endocrinol. 2021. PMID: 34545231 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

