Satellite DNA-mediated diversification of a sex-ratio meiotic drive gene family in Drosophila
- PMID: 34489561
- PMCID: PMC11188575
- DOI: 10.1038/s41559-021-01543-8
Satellite DNA-mediated diversification of a sex-ratio meiotic drive gene family in Drosophila
Abstract
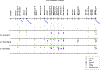
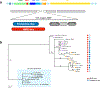
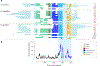
Sex chromosomes are susceptible to the evolution of selfish meiotic drive elements that bias transmission and distort progeny sex ratios. Conflict between such sex-ratio drivers and the rest of the genome can trigger evolutionary arms races resulting in genetically suppressed 'cryptic' drive systems. The Winters cryptic sex-ratio drive system of Drosophila simulans comprises a driver, Distorter on the X (Dox) and an autosomal suppressor, Not much yang, a retroduplicate of Dox that suppresses via production of endogenous small interfering RNAs (esiRNAs). Here we report that over 22 Dox-like (Dxl) sequences originated, amplified and diversified over the ~250,000-year history of the three closely related species, D. simulans, D. mauritiana and D. sechellia. The Dxl sequences encode a rapidly evolving family of protamines. Dxl copy numbers amplified by ectopic exchange among euchromatic islands of satellite DNAs on the X chromosome and separately spawned four esiRNA-producing suppressors on the autosomes. Our results reveal the genomic consequences of evolutionary arms races and highlight complex interactions among different classes of selfish DNAs.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures



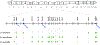


Comment in
-
A flurry of sex-ratio distorters.Nat Ecol Evol. 2021 Dec;5(12):1574-1575. doi: 10.1038/s41559-021-01601-1. Nat Ecol Evol. 2021. PMID: 34862476 No abstract available.
References
-
- Sandler L. & Novitski E. Meiotic drive as an evolutionary force. American Naturalist 91, 105–110 (1957).
-
- Lyttle TW Segregation Distorters. Annual Review of Genetics 25, 511–557 (1991). - PubMed
-
- Lyttle TW Cheaters sometimes prosper: distortion of mendelian segregation by meiotic drive. Trends in Genetics 9, 205–210 (1993). - PubMed
-
- Presgraves DC in Sperm Biology: An Evolutionary Perspective (eds Birkhead TR, Hosken DJ, & Pitnick S) (Elsevier Press, 2008).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

