Neutrophil Myeloperoxidase Derived Chlorolipid Production During Bacteria Exposure
- PMID: 34489949
- PMCID: PMC8416994
- DOI: 10.3389/fimmu.2021.701227
Neutrophil Myeloperoxidase Derived Chlorolipid Production During Bacteria Exposure
Abstract
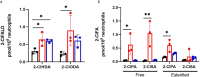
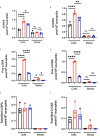
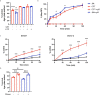
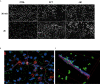
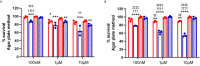
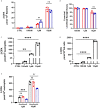
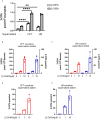
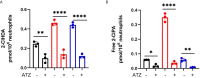
Neutrophils are the most abundant white blood cells recruited to the sites of infection and inflammation. During neutrophil activation, myeloperoxidase (MPO) is released and converts hydrogen peroxide to hypochlorous acid (HOCl). HOCl reacts with plasmalogen phospholipids to liberate 2-chlorofatty aldehyde (2-ClFALD), which is metabolized to 2-chlorofatty acid (2-ClFA). 2-ClFA and 2-ClFALD are linked with inflammatory diseases and induce endothelial dysfunction, neutrophil extracellular trap formation (NETosis) and neutrophil chemotaxis. Here we examine the neutrophil-derived chlorolipid production in the presence of pathogenic E. coli strain CFT073 and non-pathogenic E. coli strain JM109. Neutrophils cocultured with CFT073 E. coli strain and JM109 E. coli strain resulted in 2-ClFALD production. 2-ClFA was elevated only in CFT073 coculture. NETosis is more prevalent in CFT073 cocultures with neutrophils compared to JM109 cocultures. 2-ClFA and 2-ClFALD were both shown to have significant bactericidal activity, which is more severe in JM109 E. coli. 2-ClFALD metabolic capacity was 1000-fold greater in neutrophils compared to either strain of E. coli. MPO inhibition reduced chlorolipid production as well as bacterial killing capacity. These findings indicate the chlorolipid profile is different in response to these two different strains of E. coli bacteria.
Keywords: 2-chlorofatty acid; 2-chlorofatty aldehyde; E. coli; inflammation; myeloperoxidase; neutrophils; plasmalogen.
Copyright © 2021 Amunugama, Kolar and Ford.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

