Integration of Molecular Inflammatory Interactome Analyses Reveals Dynamics of Circulating Cytokines and Extracellular Vesicle Long Non-Coding RNAs and mRNAs in Heroin Addicts During Acute and Protracted Withdrawal
- PMID: 34489980
- PMCID: PMC8416766
- DOI: 10.3389/fimmu.2021.730300
Integration of Molecular Inflammatory Interactome Analyses Reveals Dynamics of Circulating Cytokines and Extracellular Vesicle Long Non-Coding RNAs and mRNAs in Heroin Addicts During Acute and Protracted Withdrawal
Abstract
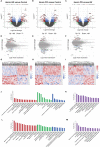
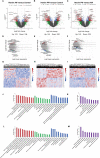
Heroin addiction and withdrawal influence multiple physiological functions, including immune responses, but the mechanism remains largely elusive. The objective of this study was to investigate the molecular inflammatory interactome, particularly the cytokines and transcriptome regulatory network in heroin addicts undergoing withdrawal, compared to healthy controls (HCs). Twenty-seven cytokines were simultaneously assessed in 41 heroin addicts, including 20 at the acute withdrawal (AW) stage and 21 at the protracted withdrawal (PW) stage, and 38 age- and gender-matched HCs. Disturbed T-helper(Th)1/Th2, Th1/Th17, and Th2/Th17 balances, characterized by reduced interleukin (IL)-2, elevated IL-4, IL-10, and IL-17A, but normal TNF-α, were present in the AW subjects. These imbalances were mostly restored to the baseline at the PW stage. However, the cytokines TNF-α, IL-2, IL-7, IL-10, and IL-17A remained dysregulated. This study also profiled exosomal long non-coding RNA (lncRNA) and mRNA in the plasma of heroin addicts, constructed co-expression gene regulation networks, and identified lncRNA-mRNA-pathway pairs specifically associated with alterations in cytokine profiles and Th1/Th2/Th17 imbalances. Altogether, a large amount of cytokine and exosomal lncRNA/mRNA expression profiling data relating to heroin withdrawal was obtained, providing a useful experimental and theoretical basis for further understanding of the pathogenic mechanisms of withdrawal symptoms in heroin addicts.
Keywords: cytokine; exosome; heroin; non-coding RNA; withdrawal.
Copyright © 2021 Zhang, Wu, Peng, Xie, Chen, Ma, Zhang, Zhou, Yang, Chen, Li, Zhang, Tian, Wang, Xu, Luo, Zhu, Kuang, Yu and Wang.
Conflict of interest statement
Author YW was employed by Echo Biotech Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Office of China National Narcotics Control Commission . China Drug Situation Report. In: The State Council China, 2018 (2019).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

