Topological Data Analysis Highlights Novel Geographical Signatures of the Human Gut Microbiome
- PMID: 34490420
- PMCID: PMC8417942
- DOI: 10.3389/frai.2021.680564
Topological Data Analysis Highlights Novel Geographical Signatures of the Human Gut Microbiome
Abstract
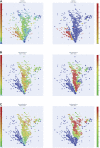
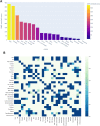
Background: There is growing interest in the connection between the gut microbiome and human health and disease. Conventional approaches to analyse microbiome data typically entail dimensionality reduction and assume linearity of the observed relationships, however, the microbiome is a highly complex ecosystem marked by non-linear relationships. In this study, we use topological data analysis (TDA) to explore differences and similarities between the gut microbiome across several countries. Methods: We used curated adult microbiome data at the genus level from the GMrepo database. The dataset contains OTU and demographical data of over 4,400 samples from 19 studies, spanning 12 countries. We analysed the data with tmap, an integrative framework for TDA specifically designed for stratification and enrichment analysis of population-based gut microbiome datasets. Results: We find associations between specific microbial genera and groups of countries. Specifically, both the USA and UK were significantly co-enriched with the proinflammatory genera Lachnoclostridium and Ruminiclostridium, while France and New Zealand were co-enriched with other, butyrate-producing, taxa of the order Clostridiales. Conclusion: The TDA approach demonstrates the overlap and distinctions of microbiome composition between and within countries. This yields unique insights into complex associations in the dataset, a finding not possible with conventional approaches. It highlights the potential utility of TDA as a complementary tool in microbiome research, particularly for large population-scale datasets, and suggests further analysis on the effects of diet and other regionally varying factors.
Keywords: global variation; gut microbiome; human microbiome; population health; topological data analysis (TDA).
Copyright © 2021 Lymberopoulos, Gentili, Alomari and Sharma.
Conflict of interest statement
Authors MA and NS are co-founders of BioCorteX Ltd. Author MA was employed by the company Rolls-Royce Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
COVID-19 severity is associated with population-level gut microbiome variations.Front Cell Infect Microbiol. 2022 Aug 23;12:963338. doi: 10.3389/fcimb.2022.963338. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36081770 Free PMC article.
-
tmap: an integrative framework based on topological data analysis for population-scale microbiome stratification and association studies.Genome Biol. 2019 Dec 23;20(1):293. doi: 10.1186/s13059-019-1871-4. Genome Biol. 2019. PMID: 31870407 Free PMC article.
-
Adjusting for age improves identification of gut microbiome alterations in multiple diseases.Elife. 2020 Mar 11;9:e50240. doi: 10.7554/eLife.50240. Elife. 2020. PMID: 32159510 Free PMC article.
-
Allogenic stem cell transplant-associated acute graft versus host disease: a computational drug discovery text mining approach using oral and gut microbiome signatures.Support Care Cancer. 2021 Apr;29(4):1765-1779. doi: 10.1007/s00520-020-05821-2. Epub 2020 Oct 22. Support Care Cancer. 2021. PMID: 33094358 Review.
-
Geography, Ethnicity or Subsistence-Specific Variations in Human Microbiome Composition and Diversity.Front Microbiol. 2017 Jun 23;8:1162. doi: 10.3389/fmicb.2017.01162. eCollection 2017. Front Microbiol. 2017. PMID: 28690602 Free PMC article. Review.
Cited by
-
COVID-19 severity is associated with population-level gut microbiome variations.Front Cell Infect Microbiol. 2022 Aug 23;12:963338. doi: 10.3389/fcimb.2022.963338. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36081770 Free PMC article.
-
Informatics for your Gut: at the Interface of Nutrition, the Microbiome, and Technology.Yearb Med Inform. 2023 Aug;32(1):89-98. doi: 10.1055/s-0043-1768723. Epub 2023 Jul 6. Yearb Med Inform. 2023. PMID: 37414029 Free PMC article. Review.
-
Gastrointestinal Manifestations of Coronavirus Disease 2019 Across the United States: A Multicenter Cohort Study.Gastro Hep Adv. 2022;1(6):909-915. doi: 10.1016/j.gastha.2022.07.002. Epub 2022 Jul 19. Gastro Hep Adv. 2022. PMID: 35874930 Free PMC article.
References
LinkOut - more resources
Full Text Sources

