Deep learning for bioimage analysis in developmental biology
- PMID: 34490888
- PMCID: PMC8451066
- DOI: 10.1242/dev.199616
Deep learning for bioimage analysis in developmental biology
Abstract
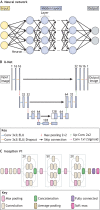
Deep learning has transformed the way large and complex image datasets can be processed, reshaping what is possible in bioimage analysis. As the complexity and size of bioimage data continues to grow, this new analysis paradigm is becoming increasingly ubiquitous. In this Review, we begin by introducing the concepts needed for beginners to understand deep learning. We then review how deep learning has impacted bioimage analysis and explore the open-source resources available to integrate it into a research project. Finally, we discuss the future of deep learning applied to cell and developmental biology. We analyze how state-of-the-art methodologies have the potential to transform our understanding of biological systems through new image-based analysis and modelling that integrate multimodal inputs in space and time.
Keywords: Bioimaging; Deep learning; Image analysis; Microscopy; Neural network.
© 2021. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures

References
-
- Adebayo, J., Gilmer, J., Muelly, M., Goodfellow, I., Hardt, M. and Kim, B. (2018). Sanity checks for saliency maps. In Proceedings of NeurIPS 2018.
-
- Barber, R. F. and Candès, E. J. (2019). A knockoff filter for high-dimensional selective inference. Annals of Statistics 47, 2504-2537. 10.1214/18-AOS1755 - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

