Catching COVID: Engineering Peptide-Modified Surface-Enhanced Raman Spectroscopy Sensors for SARS-CoV-2
- PMID: 34491043
- PMCID: PMC8442610
- DOI: 10.1021/acssensors.1c01344
Catching COVID: Engineering Peptide-Modified Surface-Enhanced Raman Spectroscopy Sensors for SARS-CoV-2
Abstract
COVID-19 remains an ongoing issue across the globe, highlighting the need for a rapid, selective, and accurate sensor for SARS-CoV-2 and its emerging variants. The chemical specificity and signal amplification of surface-enhanced Raman spectroscopy (SERS) could be advantageous for developing a quantitative assay for SARS-CoV-2 with improved speed and accuracy over current testing methods. Here, we have tackled the challenges associated with SERS detection of viruses. As viruses are large, multicomponent species, they can yield different SERS signals, but also other abundant biomolecules present in the sample can generate undesired signals. To improve selectivity in complex biological environments, we have employed peptides as capture probes for viral proteins and developed an angiotensin-converting enzyme 2 (ACE2) mimetic peptide-based SERS sensor for SARS-CoV-2. The unique vibrational signature of the spike protein bound to the peptide-modified surface is identified and used to construct a multivariate calibration model for quantification. The sensor demonstrates a 300 nM limit of detection and high selectivity in the presence of excess bovine serum albumin. This work provides the basis for designing a SERS-based assay for the detection of SARS-CoV-2 as well as engineering SERS biosensors for other viruses in the future.
Keywords: COVID-19; SARS-CoV-2; SERS; biomimetic; biosensor; peptide; virus.
Figures

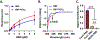



References
-
- World Health Organization. Coronavirus Disease (COVID-19) Dashboard. https://covid19.who.int.
-
- Winter L False Negatives in Quick COVID-19 Test Near 15 Percent: Study. 2020.
-
- Smith E; D G Modern Raman Spectroscopy- A Practical Approach; John Wiley & Sons, Ltd.: England, 2005.
-
- Jeanmaire DL; Van Duyne RP Surface Raman Spectroelectrochemistry. J. Electroanal. Chem. Interfacial Electrochem 1977, 84 (1), 1–20. 10.1016/s0022-0728(77)80224-6. - DOI
-
- Langer J; Jimenez de Aberasturi D; Aizpurua J; Alvarez-Puebla RA; Auguié B; Baumberg JJ; Bazan GC; Bell SEJ; Boisen A; Brolo AG; Choo J; Cialla-May D; Deckert V; Fabris L; Faulds K; García de Abajo FJ; Goodacre R; Graham D; Haes AJ; Haynes CL; Huck C; Itoh T; Käll M; Kneipp J; Kotov NA; Kuang H; Le Ru EC; Lee HK; Li J-F; Ling XY; Maier SA; Mayerhöfer T; Moskovits M; Murakoshi K; Nam J-M; Nie S; Ozaki Y; Pastoriza-Santos I; Perez-Juste J; Popp J; Pucci A; Reich S; Ren B; Schatz GC; Shegai T; Schlücker S; Tay L-L; Thomas KG; Tian Z-Q; Van Duyne RP; Vo-Dinh T; Wang Y; Willets KA; Xu C; Xu H; Xu Y; Yamamoto YS; Zhao B; Liz-Marzán LM Present and Future of Surface-Enhanced Raman Scattering. ACS Nano 2019, 14 (1), 28–117. 10.1021/acsnano.9b04224. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

