Vision, challenges and opportunities for a Plant Cell Atlas
- PMID: 34491200
- PMCID: PMC8423441
- DOI: 10.7554/eLife.66877
Vision, challenges and opportunities for a Plant Cell Atlas
Abstract
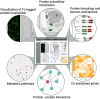
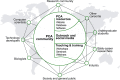
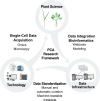
With growing populations and pressing environmental problems, future economies will be increasingly plant-based. Now is the time to reimagine plant science as a critical component of fundamental science, agriculture, environmental stewardship, energy, technology and healthcare. This effort requires a conceptual and technological framework to identify and map all cell types, and to comprehensively annotate the localization and organization of molecules at cellular and tissue levels. This framework, called the Plant Cell Atlas (PCA), will be critical for understanding and engineering plant development, physiology and environmental responses. A workshop was convened to discuss the purpose and utility of such an initiative, resulting in a roadmap that acknowledges the current knowledge gaps and technical challenges, and underscores how the PCA initiative can help to overcome them.
Keywords: 4D imaging; A. thaliana; Plant Cell Atlas; cell biology; chlamydomonas reinhardtii; location-to-function; maize; science forum; single-cell omics; translational research.
© 2021, Plant Cell Atlas Consortium et al.
Conflict of interest statement
SJ, AB, BC, NF, AF, SH, PK, ML, NP, SR, MS, PA, AA, CA, SB, JB, PD, LD, ME, SG, FG, KK, DK, SK, AM, NN, TO, MO, GP, EQ, RS, RU, JW, KV, RW, DE, KB, SR No competing interests declared
Figures




References
-
- Adamski NM, Borrill P, Brinton J, Harrington SA, Marchal C, Bentley AR, Bovill WD, Cattivelli L, Cockram J, Contreras-Moreira B, Ford B, Ghosh S, Harwood W, Hassani-Pak K, Hayta S, Hickey LT, Kanyuka K, King J, Maccaferrri M, Naamati G, Pozniak CJ, Ramirez-Gonzalez RH, Sansaloni C, Trevaskis B, Wingen LU, Wulff BB, Uauy C. A roadmap for gene functional characterisation in crops with large genomes: lessons from polyploid wheat. eLife. 2020;9:e55646. doi: 10.7554/eLife.55646. - DOI - PMC - PubMed
-
- Avise J. The hope, hype, and reality of genetic engineering: remarkable stories from agriculture, industry, medicine, and the environment. Choice. 2004;1:0283. doi: 10.5860/choice.42-0283. - DOI
-
- Avraham S, Tung C-W, Ilic K, Jaiswal P, Kellogg EA, McCouch S, Pujar A, Reiser L, Rhee SY, Sachs MM, Schaeffer M, Stein L, Stevens P, Vincent L, Zapata F, Ware D. The plant ontology database: a community resource for plant structure and developmental stages controlled vocabulary and annotations. Nucleic Acids Research. 2008;36:D449–D454. doi: 10.1093/nar/gkm908. - DOI - PMC - PubMed
-
- Balasubramanian VK, Purvine SO, Liang Y, Kelly RT, Pasa-Tolic L, Chrisler WB, Blumwald E, Stewart CN, Zhu Y, Ahkami AH. Cell-type-specific proteomics analysis of a small number of plant cells by integrating laser capture microdissection with a nanodroplet sample processing platform. Current Protocols. 2021;1:e153. doi: 10.1002/cpz1.153. - DOI - PubMed

