Phospholipase A2 enzymes represent a shared pathogenic pathway in psoriasis and pityriasis rubra pilaris
- PMID: 34491907
- PMCID: PMC8564909
- DOI: 10.1172/jci.insight.151911
Phospholipase A2 enzymes represent a shared pathogenic pathway in psoriasis and pityriasis rubra pilaris
Abstract
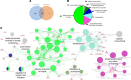
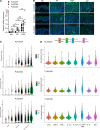
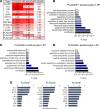
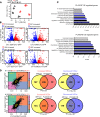
Altered epidermal differentiation along with increased keratinocyte proliferation is a characteristic feature of psoriasis and pityriasis rubra pilaris (PRP). However, despite this large degree of overlapping clinical and histologic features, the molecular signatures these skin disorders share are unknown. Using global transcriptomic profiling, we demonstrate that plaque psoriasis and PRP skin lesions have high overlap, with all differentially expressed genes in PRP relative to normal skin having complete overlap with those in psoriasis. The major common pathway shared between psoriasis and PRP involves the phospholipases PLA2G2F, PLA2G4D, and PLA2G4E, which were found to be primarily expressed in the epidermis. Gene silencing each of the 3 PLA2s led to reduction in immune responses and epidermal thickness both in vitro and in vivo in a mouse model of psoriasis, establishing their proinflammatory roles. Lipidomic analyses demonstrated that PLA2s affect mobilization of a phospholipid-eicosanoid pool, which is altered in psoriatic lesions and functions to promote immune responses in keratinocytes. Taken together, our results highlight the important role of PLA2s as regulators of epidermal barrier homeostasis and inflammation, identify PLA2s as a shared pathogenic mechanism between PRP and psoriasis, and as potential therapeutic targets for both diseases.
Keywords: Autoimmune diseases; Dermatology; Immunology; Innate immunity; Skin.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

