A systemic cell cycle block impacts stage-specific histone modification profiles during Xenopus embryogenesis
- PMID: 34491983
- PMCID: PMC8535184
- DOI: 10.1371/journal.pbio.3001377
A systemic cell cycle block impacts stage-specific histone modification profiles during Xenopus embryogenesis
Abstract
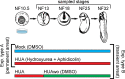
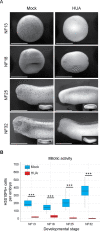
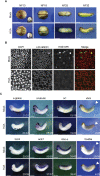
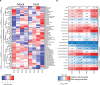
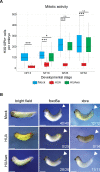
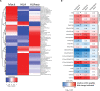
Forming an embryo from a zygote poses an apparent conflict for epigenetic regulation. On the one hand, the de novo induction of cell fate identities requires the establishment and subsequent maintenance of epigenetic information to harness developmental gene expression. On the other hand, the embryo depends on cell proliferation, and every round of DNA replication dilutes preexisting histone modifications by incorporation of new unmodified histones into chromatin. Here, we investigated the possible relationship between the propagation of epigenetic information and the developmental cell proliferation during Xenopus embryogenesis. We systemically inhibited cell proliferation during the G1/S transition in gastrula embryos and followed their development until the tadpole stage. Comparing wild-type and cell cycle-arrested embryos, we show that the inhibition of cell proliferation is principally compatible with embryo survival and cellular differentiation. In parallel, we quantified by mass spectrometry the abundance of a large set of histone modification states, which reflects the developmental maturation of the embryonic epigenome. The arrested embryos developed abnormal stage-specific histone modification profiles (HMPs), in which transcriptionally repressive histone marks were overrepresented. Embryos released from the cell cycle block during neurulation reverted toward normality on morphological, molecular, and epigenetic levels. These results suggest that the cell cycle block by HUA alters stage-specific HMPs. We propose that this influence is strong enough to control developmental decisions, specifically in cell populations that switch between resting and proliferating states such as stem cells.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Stage-specific histone modification profiles reveal global transitions in the Xenopus embryonic epigenome.PLoS One. 2011;6(7):e22548. doi: 10.1371/journal.pone.0022548. Epub 2011 Jul 22. PLoS One. 2011. PMID: 21814581 Free PMC article.
-
Developmental activation of the capability to undergo checkpoint-induced apoptosis in the early zebrafish embryo.Dev Biol. 1999 May 15;209(2):409-33. doi: 10.1006/dbio.1999.9243. Dev Biol. 1999. PMID: 10328930
-
Accumulation of histone H1(0) during early Xenopus laevis development.Exp Cell Res. 1995 Jun;218(2):586-95. doi: 10.1006/excr.1995.1196. Exp Cell Res. 1995. PMID: 7796895
-
Insights into epigenetic patterns in mammalian early embryos.Protein Cell. 2021 Jan;12(1):7-28. doi: 10.1007/s13238-020-00757-z. Epub 2020 Jul 15. Protein Cell. 2021. PMID: 32671792 Free PMC article. Review.
-
Controlling the Messenger: Regulated Translation of Maternal mRNAs in Xenopus laevis Development.Adv Exp Med Biol. 2017;953:49-82. doi: 10.1007/978-3-319-46095-6_2. Adv Exp Med Biol. 2017. PMID: 27975270 Free PMC article. Review.
Cited by
-
The histone H4K20 methyltransferase SUV4-20H1/KMT5B is required for multiciliated cell differentiation in Xenopus.Life Sci Alliance. 2023 Apr 28;6(7):e202302023. doi: 10.26508/lsa.202302023. Print 2023 Jul. Life Sci Alliance. 2023. PMID: 37116939 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

