Using test positivity and reported case rates to estimate state-level COVID-19 prevalence and seroprevalence in the United States
- PMID: 34491990
- PMCID: PMC8448371
- DOI: 10.1371/journal.pcbi.1009374
Using test positivity and reported case rates to estimate state-level COVID-19 prevalence and seroprevalence in the United States
Abstract
Accurate estimates of infection prevalence and seroprevalence are essential for evaluating and informing public health responses and vaccination coverage needed to address the ongoing spread of COVID-19 in each United States (U.S.) state. However, reliable, timely data based on representative population sampling are unavailable, and reported case and test positivity rates are highly biased. A simple data-driven Bayesian semi-empirical modeling framework was developed and used to evaluate state-level prevalence and seroprevalence of COVID-19 using daily reported cases and test positivity ratios. The model was calibrated to and validated using published state-wide seroprevalence data, and further compared against two independent data-driven mathematical models. The prevalence of undiagnosed COVID-19 infections is found to be well-approximated by a geometrically weighted average of the positivity rate and the reported case rate. Our model accurately fits state-level seroprevalence data from across the U.S. Prevalence estimates of our semi-empirical model compare favorably to those from two data-driven epidemiological models. As of December 31, 2020, we estimate nation-wide a prevalence of 1.4% [Credible Interval (CrI): 1.0%-1.9%] and a seroprevalence of 13.2% [CrI: 12.3%-14.2%], with state-level prevalence ranging from 0.2% [CrI: 0.1%-0.3%] in Hawaii to 2.8% [CrI: 1.8%-4.1%] in Tennessee, and seroprevalence from 1.5% [CrI: 1.2%-2.0%] in Vermont to 23% [CrI: 20%-28%] in New York. Cumulatively, reported cases correspond to only one third of actual infections. The use of this simple and easy-to-communicate approach to estimating COVID-19 prevalence and seroprevalence will improve the ability to make public health decisions that effectively respond to the ongoing COVID-19 pandemic.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
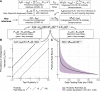

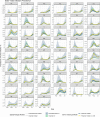
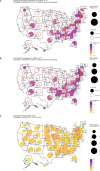
Update of
-
Using Test Positivity and Reported Case Rates to Estimate State-Level COVID-19 Prevalence and Seroprevalence in the United States.medRxiv [Preprint]. 2020 Dec 26:2020.10.07.20208504. doi: 10.1101/2020.10.07.20208504. medRxiv. 2020. Update in: PLoS Comput Biol. 2021 Sep 7;17(9):e1009374. doi: 10.1371/journal.pcbi.1009374. PMID: 33398306 Free PMC article. Updated. Preprint.
References
-
- National Academies of Sciences and Medicine E. Evaluating Data Types: A Guide for Decision Makers using Data to Understand the Extent and Spread of COVID-19 [Internet]. Washington, DC: The National Academies Press; 2020. Available from: https://www.nap.edu/catalog/25826/evaluating-data-types-a-guide-for-deci...
-
- Menachemi N, Yiannoutsos CT, Dixon BE, Duszynski TJ, Fadel WF, Wools-Kaloustian KK, et al. Population Point Prevalence of SARS-CoV-2 Infection Based on a Statewide Random Sample—Indiana, April 25–29, 2020. MMWR Morb Mortal Wkly Rep [Internet]. 2020. Jul 24 [cited 2020 Aug 28];69(29):960–4. Available from: http://www.cdc.gov/mmwr/volumes/69/wr/mm6929e1.htm?s_cid=mm6929e1_w doi: 10.15585/mmwr.mm6929e1 - DOI - PMC - PubMed
-
- Anand S, Montez-Rath M, Han J, Bozeman J, Kerschmann R, Beyer P, et al. Prevalence of SARS-CoV-2 antibodies in a large nationwide sample of patients on dialysis in the USA: a cross-sectional study. Lancet [Internet]. 2020. Oct 24 [cited 2020 Dec 14];396(10259):1335–44. Available from: doi: 10.1016/S0140-6736(20)32009-2 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

