Random forest-integrated analysis in AD and LATE brain transcriptome-wide data to identify disease-specific gene expression
- PMID: 34492068
- PMCID: PMC8423259
- DOI: 10.1371/journal.pone.0256648
Random forest-integrated analysis in AD and LATE brain transcriptome-wide data to identify disease-specific gene expression
Abstract
Alzheimer's disease (AD) is a complex neurodegenerative disorder that affects thinking, memory, and behavior. Limbic-predominant age-related TDP-43 encephalopathy (LATE) is a recently identified common neurodegenerative disease that mimics the clinical symptoms of AD. The development of drugs to prevent or treat these neurodegenerative diseases has been slow, partly because the genes associated with these diseases are incompletely understood. A notable hindrance from data analysis perspective is that, usually, the clinical samples for patients and controls are highly imbalanced, thus rendering it challenging to apply most existing machine learning algorithms to directly analyze such datasets. Meeting this data analysis challenge is critical, as more specific disease-associated gene identification may enable new insights into underlying disease-driving mechanisms and help find biomarkers and, in turn, improve prospects for effective treatment strategies. In order to detect disease-associated genes based on imbalanced transcriptome-wide data, we proposed an integrated multiple random forests (IMRF) algorithm. IMRF is effective in differentiating putative genes associated with subjects having LATE and/or AD from controls based on transcriptome-wide data, thereby enabling effective discrimination between these samples. Various forms of validations, such as cross-domain verification of our method over other datasets, improved and competitive classification performance by using identified genes, effectiveness of testing data with a classifier that is completely independent from decision trees and random forests, and relationships with prior AD and LATE studies on the genes linked to neurodegeneration, all testify to the effectiveness of IMRF in identifying genes with altered expression in LATE and/or AD. We conclude that IMRF, as an effective feature selection algorithm for imbalanced data, is promising to facilitate the development of new gene biomarkers as well as targets for effective strategies of disease prevention and treatment.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


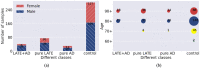

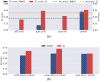
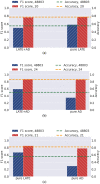



Similar articles
-
Ensemble of random forests One vs. Rest classifiers for MCI and AD prediction using ANOVA cortical and subcortical feature selection and partial least squares.J Neurosci Methods. 2018 May 15;302:47-57. doi: 10.1016/j.jneumeth.2017.12.005. Epub 2017 Dec 11. J Neurosci Methods. 2018. PMID: 29242123
-
Biomarker Extraction Based on Subspace Learning for the Prediction of Mild Cognitive Impairment Conversion.Biomed Res Int. 2021 Sep 2;2021:5531940. doi: 10.1155/2021/5531940. eCollection 2021. Biomed Res Int. 2021. PMID: 34513992 Free PMC article.
-
Classification of Alzheimer's disease and prediction of mild cognitive impairment-to-Alzheimer's conversion from structural magnetic resource imaging using feature ranking and a genetic algorithm.Comput Biol Med. 2017 Apr 1;83:109-119. doi: 10.1016/j.compbiomed.2017.02.011. Epub 2017 Feb 27. Comput Biol Med. 2017. PMID: 28260614
-
Conceptual evolution in Alzheimer's disease: implications for understanding the clinical phenotype of progressive neurodegenerative disease.J Alzheimers Dis. 2010;19(1):253-72. doi: 10.3233/JAD-2010-1237. J Alzheimers Dis. 2010. PMID: 20061643 Free PMC article. Review.
-
Antiageing strategy for neurodegenerative diseases: from mechanisms to clinical advances.Signal Transduct Target Ther. 2025 Mar 10;10(1):76. doi: 10.1038/s41392-025-02145-7. Signal Transduct Target Ther. 2025. PMID: 40059211 Free PMC article. Review.
Cited by
-
Transcriptome analysis of the Japanese eel (Anguilla japonica) during larval metamorphosis.BMC Genomics. 2024 Jun 11;25(1):585. doi: 10.1186/s12864-024-10459-z. BMC Genomics. 2024. PMID: 38862878 Free PMC article.
-
Machine Learning Approach Predicts Probability of Time to Stage-Specific Conversion of Alzheimer's Disease.J Alzheimers Dis. 2022;90(2):891-903. doi: 10.3233/JAD-220590. J Alzheimers Dis. 2022. PMID: 36189595 Free PMC article.
-
An exploratory study of high-throughput transcriptomic analysis reveals novel mRNA biomarkers for acute myocardial infarction using integrated methods.Sci Rep. 2025 Mar 11;15(1):8436. doi: 10.1038/s41598-025-92757-4. Sci Rep. 2025. PMID: 40069305 Free PMC article.
-
Deep learning algorithm reveals probabilities of stage-specific time to conversion in individuals with neurodegenerative disease LATE.Alzheimers Dement (N Y). 2022 Nov 3;8(1):e12363. doi: 10.1002/trc2.12363. eCollection 2022. Alzheimers Dement (N Y). 2022. PMID: 36348767 Free PMC article.
-
Algorithmic Stability and Generalization of an Unsupervised Feature Selection Algorithm.Adv Neural Inf Process Syst. 2021 Dec;34:19860-19875. Adv Neural Inf Process Syst. 2021. PMID: 36187051 Free PMC article.
References
-
- Chao Chen AL, Breiman L. Using Random Forest to Learn Imbalanced Data. Berkeley, California, United States: University of California; 2004.
-
- Brownlee J. Imbalanced Classification with Python: Better Metrics, Balance Skewed Classes, Cost-Sensitive Learning. 1st ed. Machine Learning Mastery; 2020.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

