The interferon landscape along the respiratory tract impacts the severity of COVID-19
- PMID: 34492226
- PMCID: PMC8373821
- DOI: 10.1016/j.cell.2021.08.016
The interferon landscape along the respiratory tract impacts the severity of COVID-19
Abstract
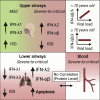
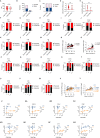
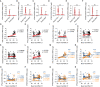
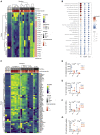
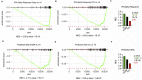
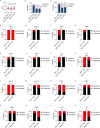
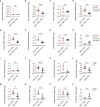
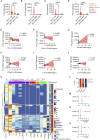
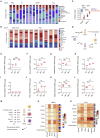
Severe coronavirus disease 2019 (COVID-19) is characterized by overproduction of immune mediators, but the role of interferons (IFNs) of the type I (IFN-I) or type III (IFN-III) families remains debated. We scrutinized the production of IFNs along the respiratory tract of COVID-19 patients and found that high levels of IFN-III, and to a lesser extent IFN-I, characterize the upper airways of patients with high viral burden but reduced disease risk or severity. Production of specific IFN-III, but not IFN-I, members denotes patients with a mild pathology and efficiently drives the transcription of genes that protect against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In contrast, compared to subjects with other infectious or noninfectious lung pathologies, IFNs are overrepresented in the lower airways of patients with severe COVID-19 that exhibit gene pathways associated with increased apoptosis and decreased proliferation. Our data demonstrate a dynamic production of IFNs in SARS-CoV-2-infected patients and show IFNs play opposing roles at distinct anatomical sites.
Keywords: COVID-19; SARS-CoV-2; Type I IFN; Type III IFN; airways; dendritic cell; epithelial cell; interferon; lung; pattern recognition receptor.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests I.Z. reports compensation for consulting services with Implicit Biosciences.
Figures

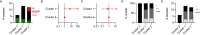




Update of
-
Severity of SARS-CoV-2 infection as a function of the interferon landscape across the respiratory tract of COVID-19 patients.bioRxiv [Preprint]. 2021 Mar 30:2021.03.30.437173. doi: 10.1101/2021.03.30.437173. bioRxiv. 2021. Update in: Cell. 2021 Sep 16;184(19):4953-4968.e16. doi: 10.1016/j.cell.2021.08.016. PMID: 33821280 Free PMC article. Updated. Preprint.
References
-
- Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., HGID Lab. NIAID-USUHS Immune Response to COVID Group. COVID Clinicians. COVID-STORM Clinicians. Imagine COVID Group. French COVID Cohort Study Group. Milieu Intérieur Consortium. CoV-Contact Cohort. Amsterdam UMC Covid-19 Biobank. COVID Human Genetic Effort Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. - PMC - PubMed
-
- Bergamaschi L., Mescia F., Turner L., Hanson A.L., Kotagiri P., Dunmore B.J., Ruffieux H., De Sa A., Huhn O., Morgan M.D., Cambridge Institute of Therapeutic Immunology and Infectious Disease-National Institute of Health Research (CITIID-NIHR) COVID BioResource Collaboration Longitudinal analysis reveals that delayed bystander CD8+ T cell activation and early immune pathology distinguish severe COVID-19 from mild disease. Immunity. 2021;54:1257–1275.e8. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

