Proximity proteomics identifies PAK4 as a component of Afadin-Nectin junctions
- PMID: 34493720
- PMCID: PMC8423818
- DOI: 10.1038/s41467-021-25011-w
Proximity proteomics identifies PAK4 as a component of Afadin-Nectin junctions
Abstract
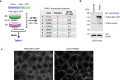
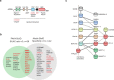
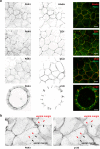
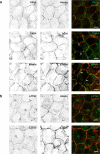
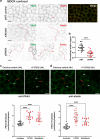
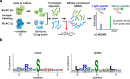
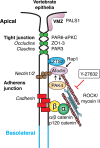
Human PAK4 is an ubiquitously expressed p21-activated kinase which acts downstream of Cdc42. Since PAK4 is enriched in cell-cell junctions, we probed the local protein environment around the kinase with a view to understanding its location and substrates. We report that U2OS cells expressing PAK4-BirA-GFP identify a subset of 27 PAK4-proximal proteins that are primarily cell-cell junction components. Afadin/AF6 showed the highest relative biotin labelling and links to the nectin family of homophilic junctional proteins. Reciprocally >50% of the PAK4-proximal proteins were identified by Afadin BioID. Co-precipitation experiments failed to identify junctional proteins, emphasizing the advantage of the BioID method. Mechanistically PAK4 depended on Afadin for its junctional localization, which is similar to the situation in Drosophila. A highly ranked PAK4-proximal protein LZTS2 was immuno-localized with Afadin at cell-cell junctions. Though PAK4 and Cdc42 are junctional, BioID analysis did not yield conventional cadherins, indicating their spatial segregation. To identify cellular PAK4 substrates we then assessed rapid changes (12') in phospho-proteome after treatment with two PAK inhibitors. Among the PAK4-proximal junctional proteins seventeen PAK4 sites were identified. We anticipate mammalian group II PAKs are selective for the Afadin/nectin sub-compartment, with a demonstrably distinct localization from tight and cadherin junctions.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

