Genotypes of informative loci from 1000 Genomes data allude evolution and mixing of human populations
- PMID: 34493766
- PMCID: PMC8423758
- DOI: 10.1038/s41598-021-97129-2
Genotypes of informative loci from 1000 Genomes data allude evolution and mixing of human populations
Abstract
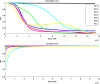
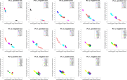
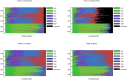
Principal Component Analysis (PCA) projects high-dimensional genotype data into a few components that discern populations. Ancestry Informative Markers (AIMs) are a small subset of SNPs capable of distinguishing populations. We integrate these two approaches by proposing an algorithm to identify necessary informative loci whose removal from the data deteriorates the PCA structure. Unlike classical AIMs, necessary informative loci densely cover the genome, hence can illuminate the evolution and mixing history of populations. We conduct a comprehensive analysis to the genotype data of the 1000 Genomes Project using necessary informative loci. Projections along the top seven principal components demarcate populations at distinct geographic levels. Millions of necessary informative loci along each PC are identified. Population identities along each PC are approximately determined by weighted sums of minor (or major) alleles over the informative loci. Variations of allele frequencies are aligned with the history and direction of population evolution. The population distribution of projections along the top three PCs is recapitulated by a simple demographic model based on several waves of founder population separation and mixing. Informative loci possess locational concentration in the genome and functional enrichment. Genes at two hot spots encompassing dense PC 7 informative loci exhibit differential expressions among European populations. The mosaic of local ancestry in the genome of a mixed descendant from multiple populations can be inferred from partial PCA projections of informative loci. Finally, informative loci derived from the 1000 Genomes data well predict the projections of an independent genotype data of South Asians. These results demonstrate the utility and relevance of informative loci to investigate human evolution.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Cavalli-Sforza LL, et al. The History and Geography of Human Genes. Princeton University Press; 1994.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

