Nanopore sequencing and de novo assembly of a misidentified Camelpox vaccine reveals putative epigenetic modifications and alternate protein signal peptides
- PMID: 34493784
- PMCID: PMC8423768
- DOI: 10.1038/s41598-021-97158-x
Nanopore sequencing and de novo assembly of a misidentified Camelpox vaccine reveals putative epigenetic modifications and alternate protein signal peptides
Abstract
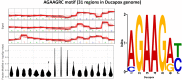
DNA viruses can exploit host cellular epigenetic processes to their advantage; however, the epigenome status of most DNA viruses remains undetermined. Third generation sequencing technologies allow for the identification of modified nucleotides from sequencing experiments without specialized sample preparation, permitting the detection of non-canonical epigenetic modifications that may distinguish viral nucleic acid from that of their host, thus identifying attractive targets for advanced therapeutics and diagnostics. We present a novel nanopore de novo assembly pipeline used to assemble a misidentified Camelpox vaccine. Two confirmed deletions of this vaccine strain in comparison to the closely related Vaccinia virus strain modified vaccinia Ankara make it one of the smallest non-vector derived orthopoxvirus genomes to be reported. Annotation of the assembly revealed a previously unreported signal peptide at the start of protein A38 and several predicted signal peptides that were found to differ from those previously described. Putative epigenetic modifications around various motifs have been identified and the assembly confirmed previous work showing the vaccine genome to most closely resemble that of Vaccinia virus strain Modified Vaccinia Ankara. The pipeline may be used for other DNA viruses, increasing the understanding of DNA virus evolution, virulence, host preference, and epigenomics.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Fenner, F., Henderson, D.A., Arita, I., Jezek, Z. & Ladnyi, I.D. Smallpox and its eradication. Geneva: World Health Organization; 1988. [March 14, 2003]. p. 1460. Reference out-of-print. See the World Health Organization, Communicable Disease Surveillance and Response Web site. www.who.int/emc/diseases/smallpox/smallpoxeradication.html.
-
- Jenner, E. An inquiry into the causes and effects of the variole vaccinae, a disease discovered in some of the Western Counties of England, Particularly Gloucestershire and Known by the Name of the cow‐pox. London: Sampson Low, 1798.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

