Lymphoid-specific helicase in epigenetics, DNA repair and cancer
- PMID: 34493821
- PMCID: PMC8770686
- DOI: 10.1038/s41416-021-01543-2
Lymphoid-specific helicase in epigenetics, DNA repair and cancer
Abstract
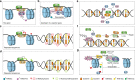
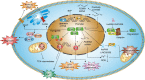
Lymphoid-specific helicase (LSH) is a member of the SNF2 helicase family of chromatin-remodelling proteins. Dysfunctions or mutations in LSH causes an autosomal recessive disease known as immunodeficiency-centromeric instability-facial anomaly (ICF) syndrome. Interestingly, LSH participates in various aspects of epigenetic regulation, including nucleosome remodelling, DNA methylation, histone modifications and heterochromatin formation. Further, LSH plays a crucial role during DNA-damage repair, specifically during double-strand break (DSB) repair, since murine LSH was shown to be essential for non-homologous end joining (NHEJ) and homologous recombination (HR). Accordingly, overexpression of LSH drives tumorigenesis and malignancy. On the other hand, LSH homologs stabilise the genome. Thus, LSH might be implemented as a biomarker for various cancer types and potential target molecule to develop therapeutic strategies against them. In this review, we focus on the role of LSH in orchestrating chromatin rearrangements, such as DNA methylation and histone modifications, as well as in DNA-damage repair. Changes in chromatin structure may facilitate gene expression signatures that cause malignant transformation. We summarise recent findings of LSH in cancers and raise critical open questions for further studies.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests. This manuscript has been read and approved by all the authors, and has not been submitted to or is not under consideration for publication elsewhere.
Figures


References
-
- Cavalli G, Heard E. Advances in epigenetics link genetics to the environment and disease. Nature. 2019;571:489–99. - PubMed
-
- Dobersch, S, Rubio, K, Barreto, G. Pioneer factors and architectural proteins mediating embryonic expression signatures in cancer. Trends Mol Med. 2019. 10.1016/j.molmed.2019.01.008. - PubMed
-
- Singh AK, Mueller-Planitz F. Nucleosome positioning and spacing: from mechanism to function. J Mol Biol. 2021;433:166847. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

