Genomic and phenotypic insights from an atlas of genetic effects on DNA methylation
- PMID: 34493871
- PMCID: PMC7612069
- DOI: 10.1038/s41588-021-00923-x
Genomic and phenotypic insights from an atlas of genetic effects on DNA methylation
Abstract
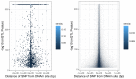
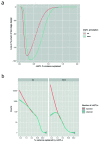
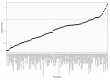
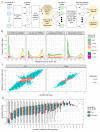
Characterizing genetic influences on DNA methylation (DNAm) provides an opportunity to understand mechanisms underpinning gene regulation and disease. In the present study, we describe results of DNAm quantitative trait locus (mQTL) analyses on 32,851 participants, identifying genetic variants associated with DNAm at 420,509 DNAm sites in blood. We present a database of >270,000 independent mQTLs, of which 8.5% comprise long-range (trans) associations. Identified mQTL associations explain 15-17% of the additive genetic variance of DNAm. We show that the genetic architecture of DNAm levels is highly polygenic. Using shared genetic control between distal DNAm sites, we constructed networks, identifying 405 discrete genomic communities enriched for genomic annotations and complex traits. Shared genetic variants are associated with both DNAm levels and complex diseases, but only in a minority of cases do these associations reflect causal relationships from DNAm to trait or vice versa, indicating a more complex genotype-phenotype map than previously anticipated.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
T.R.G receives funding from GlaxoSmithKline and Biogen for unrelated research. Other authors declare no competing interests.
Figures








References
-
- Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465:721–7. - PubMed
-
- Kerkel K, et al. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet. 2008;40:904–8. - PubMed
-
- Schadt EE, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_UU_00007/10/MRC_/Medical Research Council/United Kingdom
- MC_PC_19009/MRC_/Medical Research Council/United Kingdom
- ALCHALABI-DOBSON/APR14/829-791/MNDA_/Motor Neurone Disease Association/United Kingdom
- CS/13/1/30327/BHF_/British Heart Foundation/United Kingdom
- MC_UU_00011/5/MRC_/Medical Research Council/United Kingdom
- 529051021/ZONMW_/ZonMw/Netherlands
- MR/M008924/1/MRC_/Medical Research Council/United Kingdom
- G0600974/MRC_/Medical Research Council/United Kingdom
- G1002190/MRC_/Medical Research Council/United Kingdom
- ALCHALABI-TALBOT/APR14/926-794/MNDA_/Motor Neurone Disease Association/United Kingdom
- 208806/WT_/Wellcome Trust/United Kingdom
- MC_UU_00006/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_00006/2/MRC_/Medical Research Council/United Kingdom
- G9815508/MRC_/Medical Research Council/United Kingdom
- MR/L501529/1/MRC_/Medical Research Council/United Kingdom
- 204979/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- CS/15/6/31468/BHF_/British Heart Foundation/United Kingdom
- MR/N024397/1/MRC_/Medical Research Council/United Kingdom
- CS/16/4/32482/BHF_/British Heart Foundation/United Kingdom
- G1001357/MRC_/Medical Research Council/United Kingdom
- MC_PC_21038/MRC_/Medical Research Council/United Kingdom
- MR/K013807/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_12015/2/MRC_/Medical Research Council/United Kingdom
- MC_PC_15018/MRC_/Medical Research Council/United Kingdom
- MC_UU_00011/4/MRC_/Medical Research Council/United Kingdom
- C18281/A191169/Cancer Research UK (CRUK)
- MR/R024804/1/MRC_/Medical Research Council/United Kingdom
- BB/S020845/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 208806/Z/17/Z/WT_/Wellcome Trust/United Kingdom
- MR/R005176/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_00011/1/MRC_/Medical Research Council/United Kingdom
- A19169/CRUK_/Cancer Research UK/United Kingdom
- 204979/WT_/Wellcome Trust/United Kingdom
- MC_UU_12013/8/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources

