This is a preprint.
Enzymatic Beacons for Specific Sensing of Dilute Nucleic Acid and Potential Utility for SARS-CoV-2 Detection
- PMID: 34494022
- PMCID: PMC8423218
- DOI: 10.1101/2021.08.30.458287
Enzymatic Beacons for Specific Sensing of Dilute Nucleic Acid and Potential Utility for SARS-CoV-2 Detection
Update in
-
Enzymatic Beacons for Specific Sensing of Dilute Nucleic Acid.Chembiochem. 2022 Feb 16;23(4):e202100594. doi: 10.1002/cbic.202100594. Epub 2021 Dec 29. Chembiochem. 2022. PMID: 34890095 Free PMC article.
Abstract
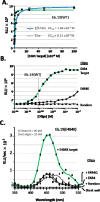
Enzymatic beacons, or E-beacons, are 1:1 bioconjugates of the nanoluciferase enzyme linked covalently at its C-terminus to hairpin forming DNA oligonucleotides equipped with a dark quencher. We prepared E-beacons biocatalytically using the promiscuous "hedgehog" protein-cholesterol ligase, HhC. Instead of cholesterol, HhC attached nanoluciferase site-specifically to mono-sterylated hairpin DNA, prepared in high yield by solid phase synthesis. We tested three potential E-beacon dark quenchers: Iowa Black, Onyx-A, and dabcyl. Prototype E-beacon carrying each of those quenchers provided sequence-specific nucleic acid sensing through turn-on bioluminescence. For practical application, we prepared dabcyl-quenched E-beacons for potential use in detecting the COVID-19 virus, SARS-CoV-2. Targeting the E484 codon of the SARS-CoV-2 Spike protein, E-beacons (80 × 10 -12 M) reported wild-type SARS-CoV-2 nucleic acid at ≥1 × 10 -9 M with increased bioluminescence of 8-fold. E-beacon prepared for the E484K variant of SARS-CoV-2 functioned with similar sensitivity. These E-beacons could discriminate their complementary target from nucleic acid encoding the E484Q mutation of the SARS-CoV-2 Kappa variant. Along with specificity, detection sensitivity with E-beacons is two to three orders of magnitude better than synthetic molecular beacons, rivaling the most sensitive nucleic acid detection agents reported to date.
Figures


References
-
- Corman V. M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D. K., Bleicker T., Brunink S., Schneider J., Schmidt M. L., Mulders D. G., Haagmans B. L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J. L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M. P., and Drosten C. (2020) Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR, Euro Surveill 25. - PMC - PubMed
-
- Arons M. M., Hatfield K. M., Reddy S. C., Kimball A., James A., Jacobs J. R., Taylor J., Spicer K., Bardossy A. C., Oakley L. P., Tanwar S., Dyal J. W., Harney J., Chisty Z., Bell J. M., Methner M., Paul P., Carlson C. M., McLaughlin H. P., Thornburg N., Tong S., Tamin A., Tao Y., Uehara A., Harcourt J., Clark S., Brostrom-Smith C., Page L. C., Kay M., Lewis J., Montgomery P., Stone N. D., Clark T. A., Honein M. A., Duchin J. S., Jernigan J. A., Public H.-S., King C., and Team C. C.-I. (2020) Presymptomatic SARS-CoV-2 Infections and Transmission in a Skilled Nursing Facility, N Engl J Med 382, 2081–2090. - PMC - PubMed
-
- Hosseini A., Pandey R., Osman E., Victorious A., Li F., Didar T., and Soleymani L. (2020) Roadmap to the Bioanalytical Testing of COVID-19: From Sample Collection to Disease Surveillance, ACS Sens 5, 3328–3345. - PubMed
-
- Marras S. A., Kramer F. R., and Tyagi S. (1999) Multiplex detection of single-nucleotide variations using molecular beacons, Genet Anal 14, 151–156. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
