VC@Scale: Scalable and high-performance variant calling on cluster environments
- PMID: 34494101
- PMCID: PMC8424057
- DOI: 10.1093/gigascience/giab057
VC@Scale: Scalable and high-performance variant calling on cluster environments
Abstract
Background: Recently many new deep learning-based variant-calling methods like DeepVariant have emerged as more accurate compared with conventional variant-calling algorithms such as GATK HaplotypeCaller, Sterlka2, and Freebayes albeit at higher computational costs. Therefore, there is a need for more scalable and higher performance workflows of these deep learning methods. Almost all existing cluster-scaled variant-calling workflows that use Apache Spark/Hadoop as big data frameworks loosely integrate existing single-node pre-processing and variant-calling applications. Using Apache Spark just for distributing/scheduling data among loosely coupled applications or using I/O-based storage for storing the output of intermediate applications does not exploit the full benefit of Apache Spark in-memory processing. To achieve this, we propose a native Spark-based workflow that uses Python and Apache Arrow to enable efficient transfer of data between different workflow stages. This benefits from the ease of programmability of Python and the high efficiency of Arrow's columnar in-memory data transformations.
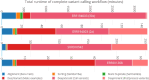
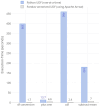
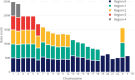
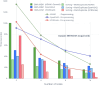
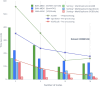
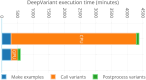
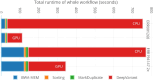
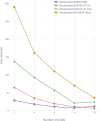
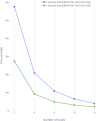
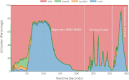
Results: Here we present a scalable, parallel, and efficient implementation of next-generation sequencing data pre-processing and variant-calling workflows. Our design tightly integrates most pre-processing workflow stages, using Spark built-in functions to sort reads by coordinates and mark duplicates efficiently. Our approach outperforms state-of-the-art implementations by >2 times for the pre-processing stages, creating a scalable and high-performance solution for DeepVariant for both CPU-only and CPU + GPU clusters.
Conclusions: We show the feasibility and easy scalability of our approach to achieve high performance and efficient resource utilization for variant-calling analysis on high-performance computing clusters using the standardized Apache Arrow data representations. All codes, scripts, and configurations used to run our implementations are publicly available and open sourced; see https://github.com/abs-tudelft/variant-calling-at-scale.
Keywords: Apache Arrow; Apache Spark; BWA-MEM; DeepVariant; MarkDuplicate; sorting; whole-genome sequencing.
© The Author(s) 2021. Published by Oxford University Press GigaScience.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Gropp W, Lusk E. Fault tolerance in message passing interface programs. Int J High Perform Comput Appl. 2004;18(3):363–72.
-
- Cappello F, Al G, Gropp W, et al. Toward exascale resilience: 2014 update. Supercomput Front Innov. 2014;1(1):5–28.
-
- Apache Apache Hadoop. 2019. https://hadoop.apache.org/. Accessed 2 April 2019.
-
- Apache. Apache Spark: Lightning-fast unified analytics engine. 2019. https://spark.apache.org/. Accessed 2 April 2019.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

