The interplay of RNA:DNA hybrid structure and G-quadruplexes determines the outcome of R-loop-replisome collisions
- PMID: 34494544
- PMCID: PMC8479836
- DOI: 10.7554/eLife.72286
The interplay of RNA:DNA hybrid structure and G-quadruplexes determines the outcome of R-loop-replisome collisions
Abstract
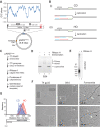
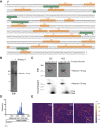
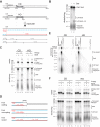
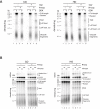
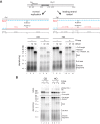
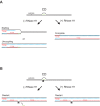
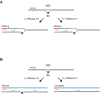
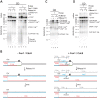
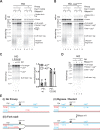
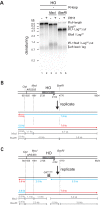
R-loops are a major source of genome instability associated with transcription-induced replication stress. However, how R-loops inherently impact replication fork progression is not understood. Here, we characterize R-loop-replisome collisions using a fully reconstituted eukaryotic DNA replication system. We find that RNA:DNA hybrids and G-quadruplexes at both co-directional and head-on R-loops can impact fork progression by inducing fork stalling, uncoupling of leading strand synthesis from replisome progression, and nascent strand gaps. RNase H1 and Pif1 suppress replication defects by resolving RNA:DNA hybrids and G-quadruplexes, respectively. We also identify an intrinsic capacity of replisomes to maintain fork progression at certain R-loops by unwinding RNA:DNA hybrids, repriming leading strand synthesis downstream of G-quadruplexes, or utilizing R-loop transcripts to prime leading strand restart during co-directional R-loop-replisome collisions. Collectively, the data demonstrates that the outcome of R-loop-replisome collisions is modulated by R-loop structure, providing a mechanistic basis for the distinction of deleterious from non-deleterious R-loops.
Keywords: DNA replication; G-quadruplex; Pif1; R-loop; RNase H; S. cerevisiae; biochemistry; chemical biology; chromosomes; gene expression; replisome; transcription-replication conflict.
© 2021, Kumar et al.
Conflict of interest statement
CK, SB, JG, DR No competing interests declared
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

