Tumor-initiating stem cell shapes its microenvironment into an immunosuppressive barrier and pro-tumorigenic niche
- PMID: 34496236
- PMCID: PMC8451448
- DOI: 10.1016/j.celrep.2021.109674
Tumor-initiating stem cell shapes its microenvironment into an immunosuppressive barrier and pro-tumorigenic niche
Abstract
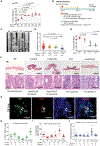
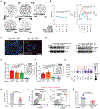
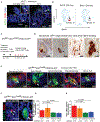
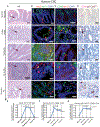
Tumor-initiating stem cells (TSCs) are critical for drug resistance and immune escape. However, the mutual regulations between TSC and tumor microenvironment (TME) remain unclear. Using DNA-label retaining, single-cell RNA sequencing (scRNA-seq), and other approaches, we investigated intestinal adenoma in response to chemoradiotherapy (CRT), thus identifying therapy-resistant TSCs (TrTSCs). We find bidirectional crosstalk between TSCs and TME using CellPhoneDB analysis. An intriguing finding is that TSCs shape TME into a landscape that favors TSCs for immunosuppression and propagation. Using adenoma-organoid co-cultures, niche-cell depletion, and lineaging tracing, we characterize a functional role of cyclooxygenase-2 (Cox-2)-dependent signaling, predominantly occurring between tumor-associated monocytes and macrophages (TAMMs) and TrTSCs. We show that TAMMs promote TrTSC proliferation through prostaglandin E2 (PGE2)-PTGER4(EP4) signaling, which enhances β-catenin activity via AKT phosphorylation. Thus, our study shows that the bidirectional crosstalk between TrTSC and TME results in a pro-tumorigenic and immunosuppressive contexture.
Keywords: C5AR1; CD74; EP4; MIF; Pge2; RPS19; immune barrier; mciroenvironment; niche; resistance; stem cell.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Aoki T, Frȍsen J, Fukuda M, Bando K, Shioi G, Tsuji K, Ollikainen E, Nozaki K, Laakkonen J, and Narumiya S (2017). Prostaglandin E2-EP2-NF-κB signaling in macrophages as a potential therapeutic target for intracranial aneurysms. Sci. Signal 10, eaah6037. - PubMed
-
- Ayyaz A, Kumar S, Sangiorgi B, Ghoshal B, Gosio J, Ouladan S, Fink M, Barutcu S, Trcka D, Shen J, et al. (2019). Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature 569, 121–125. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

