Proximity labeling and other novel mass spectrometric approaches for spatiotemporal protein dynamics
- PMID: 34496693
- PMCID: PMC8650568
- DOI: 10.1080/14789450.2021.1976149
Proximity labeling and other novel mass spectrometric approaches for spatiotemporal protein dynamics
Abstract
Background: Proteins are highly dynamic and their biological function is controlled by not only temporal abundance changes but also via regulated protein-protein interaction networks, which respond to internal and external perturbations. A wealth of novel analytical reagents and workflows allow studying spatiotemporal protein environments with great granularity while maintaining high throughput and ease of analysis.
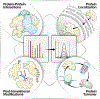
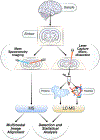
Areas covered: We review technology advances for measuring protein-protein proximity interactions with an emphasis on proximity labeling, and briefly summarize other spatiotemporal approaches including protein localization, and their dynamic changes over time, specifically in human cells and mammalian tissues. We focus especially on novel technologies and workflows emerging within the past 5 years. This includes enrichment-based techniques (proximity labeling and crosslinking), separation-based techniques (organelle fractionation and size exclusion chromatography), and finally sorting-based techniques (laser capture microdissection and mass spectrometry imaging).
Expert opinion: Spatiotemporal proteomics is a key step in assessing biological complexity, understanding refined regulatory mechanisms, and forming protein complexes and networks. Studying protein dynamics across space and time holds promise for gaining deep insights into how protein networks may be perturbed during disease and aging processes, and offer potential avenues for therapeutic interventions, drug discovery, and biomarker development.
Keywords: Chemical crosslinking; mass spectrometry; protein interactome; protein networks; protein turnover; proximity labeling.
Conflict of interest statement
Declaration of interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Figures

Similar articles
-
Biotinylation-based proximity labelling proteomics: basics, applications and technical considerations.J Biochem. 2021 Dec 28;170(5):569-576. doi: 10.1093/jb/mvab123. J Biochem. 2021. PMID: 34752609 Review.
-
Recent Advances in Mass Spectrometry-Based Protein Interactome Studies.Mol Cell Proteomics. 2025 Jan;24(1):100887. doi: 10.1016/j.mcpro.2024.100887. Epub 2024 Nov 27. Mol Cell Proteomics. 2025. PMID: 39608603 Free PMC article. Review.
-
Application and progress of proximity labeling technology in the study of protein-protein interactions.Yi Chuan. 2025 Jun;47(6):625-635. doi: 10.16288/j.yczz.24-307. Yi Chuan. 2025. PMID: 40528466 Review.
-
Mass spectrometry-based methods for analysing the mitochondrial interactome in mammalian cells.J Biochem. 2020 Mar 1;167(3):225-231. doi: 10.1093/jb/mvz090. J Biochem. 2020. PMID: 31647556 Free PMC article. Review.
-
[Recent advances in glycopeptide enrichment and mass spectrometry data interpretation approaches for glycoproteomics analyses].Se Pu. 2021 Oct;39(10):1045-1054. doi: 10.3724/SP.J.1123.2021.06011. Se Pu. 2021. PMID: 34505426 Free PMC article. Review. Chinese.
Cited by
-
Endothelial cilia dysfunction in pathogenesis of hereditary hemorrhagic telangiectasia.Front Cell Dev Biol. 2022 Nov 10;10:1037453. doi: 10.3389/fcell.2022.1037453. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36438574 Free PMC article.
-
Development and Evaluation of a Robust Sandwich Immunoassay System Detecting Serum WFA-Reactive IgA1 for Diagnosis of IgA Nephropathy.Int J Mol Sci. 2022 May 5;23(9):5165. doi: 10.3390/ijms23095165. Int J Mol Sci. 2022. PMID: 35563555 Free PMC article.
-
ACKR3 Proximity Labeling Identifies Novel G protein- and β-arrestin-independent GPCR Interacting Proteins.bioRxiv [Preprint]. 2024 Jan 28:2024.01.27.577545. doi: 10.1101/2024.01.27.577545. bioRxiv. 2024. PMID: 38410489 Free PMC article. Preprint.
-
Recent Advancements in Subcellular Proteomics: Growing Impact of Organellar Protein Niches on the Understanding of Cell Biology.J Proteome Res. 2024 Aug 2;23(8):2700-2722. doi: 10.1021/acs.jproteome.3c00839. Epub 2024 Mar 7. J Proteome Res. 2024. PMID: 38451675 Free PMC article. Review.
-
Recent Advances in the Prediction of Subcellular Localization of Proteins and Related Topics.Front Bioinform. 2022 May 19;2:910531. doi: 10.3389/fbinf.2022.910531. eCollection 2022. Front Bioinform. 2022. PMID: 36304291 Free PMC article. Review.
References
-
- Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001. 2001/02/01;409(6822):860–921. - PubMed
-
- Cohen AA, Geva-Zatorsky N, Eden E, et al. Dynamic Proteomics of Individual Cancer Cells in Response to a Drug. Science. 2008. 2008/12/05/;322(5907):1511–1516. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
