A novel automated image analysis pipeline for quantifying morphological changes to the endoplasmic reticulum in cultured human cells
- PMID: 34496765
- PMCID: PMC8425006
- DOI: 10.1186/s12859-021-04334-x
A novel automated image analysis pipeline for quantifying morphological changes to the endoplasmic reticulum in cultured human cells
Abstract
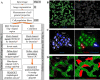
Background: In mammalian cells the endoplasmic reticulum (ER) comprises a highly complex reticular morphology that is spread throughout the cytoplasm. This organelle is of particular interest to biologists, as its dysfunction is associated with numerous diseases, which often manifest themselves as changes to the structure and organisation of the reticular network. Due to its complex morphology, image analysis methods to quantitatively describe this organelle, and importantly any changes to it, are lacking.
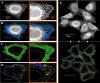
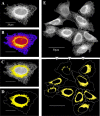
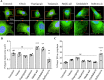
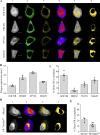
Results: In this work we detail a methodological approach that utilises automated high-content screening microscopy to capture images of cells fluorescently-labelled for various ER markers, followed by their quantitative analysis. We propose that two key metrics, namely the area of dense ER and the area of polygonal regions in between the reticular elements, together provide a basis for measuring the quantities of rough and smooth ER, respectively. We demonstrate that a number of different pharmacological perturbations to the ER can be quantitatively measured and compared in our automated image analysis pipeline. Furthermore, we show that this method can be implemented in both commercial and open-access image analysis software with comparable results.
Conclusions: We propose that this method has the potential to be applied in the context of large-scale genetic and chemical perturbations to assess the organisation of the ER in adherent cell cultures.
Keywords: Automated image analysis; ER function; Endoplasmic reticulum morphology; High-content imaging.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

