PUResNet: prediction of protein-ligand binding sites using deep residual neural network
- PMID: 34496970
- PMCID: PMC8424938
- DOI: 10.1186/s13321-021-00547-7
PUResNet: prediction of protein-ligand binding sites using deep residual neural network
Abstract
Background: Predicting protein-ligand binding sites is a fundamental step in understanding the functional characteristics of proteins, which plays a vital role in elucidating different biological functions and is a crucial step in drug discovery. A protein exhibits its true nature after binding to its interacting molecule known as a ligand that binds only in the favorable binding site of the protein structure. Different computational methods exploiting the features of proteins have been developed to identify the binding sites in the protein structure, but none seems to provide promising results, and therefore, further investigation is required.
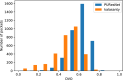
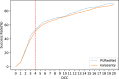
Results: In this study, we present a deep learning model PUResNet and a novel data cleaning process based on structural similarity for predicting protein-ligand binding sites. From the whole scPDB (an annotated database of druggable binding sites extracted from the Protein DataBank) database, 5020 protein structures were selected to address this problem, which were used to train PUResNet. With this, we achieved better and justifiable performance than the existing methods while evaluating two independent sets using distance, volume and proportion metrics.
Keywords: Binding site prediction; Convolutional neural network; Data cleaning; Deep residual network; Ligand binding sites.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures













References
-
- Nelson DL. Lehninger principles of biochemistry. 4. New York: W.H. Freeman; 2005.
Grants and funding
LinkOut - more resources
Full Text Sources

