Advancements in detection of SARS-CoV-2 infection for confronting COVID-19 pandemics
- PMID: 34497366
- PMCID: PMC8424153
- DOI: 10.1038/s41374-021-00663-w
Advancements in detection of SARS-CoV-2 infection for confronting COVID-19 pandemics
Abstract
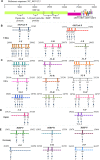
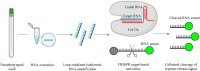
As one of the major approaches in combating the COVID-19 pandemics, the availability of specific and reliable assays for the SARS-CoV-2 viral genome and its proteins is essential to identify the infection in suspected populations, make diagnoses in symptomatic or asymptomatic individuals, and determine clearance of the virus after the infection. For these purposes, use of the quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) for detection of the viral nucleic acid remains the most valuable in terms of its specificity, fast turn-around, high-throughput capacity, and reliability. It is critical to update the sequences of primers and probes to ensure the detection of newly emerged variants. Various assays for increased levels of IgG or IgM antibodies are available for detecting ongoing or past infection, vaccination responses, and persistence and for identifying high titers of neutralizing antibodies in recovered individuals. Viral genome sequencing is increasingly used for tracing infectious sources, monitoring mutations, and subtype classification and is less valuable in diagnosis because of its capacity and high cost. Nanopore target sequencing with portable options is available for a quick process for sequencing data. Emerging CRISPR-Cas-based assays, such as SHERLOCK and AIOD-CRISPR, for viral genome detection may offer options for prompt and point-of-care detection. Moreover, aptamer-based probes may be multifaceted for developing portable and high-throughput assays with fluorescent or chemiluminescent probes for viral proteins. In conclusion, assays are available for viral genome and protein detection, and the selection of specific assays depends on the purposes of prevention, diagnosis and pandemic control, or monitoring of vaccination efficacy.
© 2021. The Author(s), under exclusive licence to United States and Canadian Academy of Pathology.
Figures

References
-
- Chan JF, et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020;58:e00310–e00320. PMCID 7180250 PubMed PMID: 32132196. - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

