Cells of the human intestinal tract mapped across space and time
- PMID: 34497389
- PMCID: PMC8426186
- DOI: 10.1038/s41586-021-03852-1
Cells of the human intestinal tract mapped across space and time
Abstract
The cellular landscape of the human intestinal tract is dynamic throughout life, developing in utero and changing in response to functional requirements and environmental exposures. Here, to comprehensively map cell lineages, we use single-cell RNA sequencing and antigen receptor analysis of almost half a million cells from up to 5 anatomical regions in the developing and up to 11 distinct anatomical regions in the healthy paediatric and adult human gut. This reveals the existence of transcriptionally distinct BEST4 epithelial cells throughout the human intestinal tract. Furthermore, we implicate IgG sensing as a function of intestinal tuft cells. We describe neural cell populations in the developing enteric nervous system, and predict cell-type-specific expression of genes associated with Hirschsprung's disease. Finally, using a systems approach, we identify key cell players that drive the formation of secondary lymphoid tissue in early human development. We show that these programs are adopted in inflammatory bowel disease to recruit and retain immune cells at the site of inflammation. This catalogue of intestinal cells will provide new insights into cellular programs in development, homeostasis and disease.
© 2021. The Author(s).
Conflict of interest statement
In the past three years, S.A.T. has consulted for or been a member of scientific advisory boards at Roche, Qiagen, Genentech, Biogen, GlaxoSmithKline and ForeSite Labs. The remaining authors declare no competing interests.
Figures
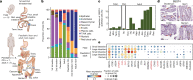





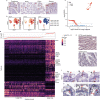

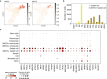










Comment in
-
Mapping the human gut.Nat Rev Gastroenterol Hepatol. 2021 Nov;18(11):747. doi: 10.1038/s41575-021-00530-5. Nat Rev Gastroenterol Hepatol. 2021. PMID: 34584255 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
- MC_PC_17230/MRC_/Medical Research Council/United Kingdom
- MR/R006237/1/WT_/Wellcome Trust/United Kingdom
- 203151/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- ERC_/European Research Council/International
- MR/T001917/1/MRC_/Medical Research Council/United Kingdom
- 220268/Z/20/Z/WT_/Wellcome Trust/United Kingdom
- MR/R006237/1/MRC_/Medical Research Council/United Kingdom
- 206194/WT_/Wellcome Trust/United Kingdom
- 16567/CRUK_/Cancer Research UK/United Kingdom
- 213555/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- MR/P028160/1/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- MR/S035842/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

