Inter-cellular CRISPR screens reveal regulators of cancer cell phagocytosis
- PMID: 34497417
- PMCID: PMC9419706
- DOI: 10.1038/s41586-021-03879-4
Inter-cellular CRISPR screens reveal regulators of cancer cell phagocytosis
Abstract
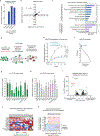
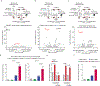
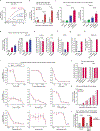
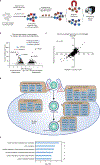
Monoclonal antibody therapies targeting tumour antigens drive cancer cell elimination in large part by triggering macrophage phagocytosis of cancer cells1-7. However, cancer cells evade phagocytosis using mechanisms that are incompletely understood. Here we develop a platform for unbiased identification of factors that impede antibody-dependent cellular phagocytosis (ADCP) using complementary genome-wide CRISPR knockout and overexpression screens in both cancer cells and macrophages. In cancer cells, beyond known factors such as CD47, we identify many regulators of susceptibility to ADCP, including the poorly characterized enzyme adipocyte plasma membrane-associated protein (APMAP). We find that loss of APMAP synergizes with tumour antigen-targeting monoclonal antibodies and/or CD47-blocking monoclonal antibodies to drive markedly increased phagocytosis across a wide range of cancer cell types, including those that are otherwise resistant to ADCP. Additionally, we show that APMAP loss synergizes with several different tumour-targeting monoclonal antibodies to inhibit tumour growth in mice. Using genome-wide counterscreens in macrophages, we find that the G-protein-coupled receptor GPR84 mediates enhanced phagocytosis of APMAP-deficient cancer cells. This work reveals a cancer-intrinsic regulator of susceptibility to antibody-driven phagocytosis and, more broadly, expands our knowledge of the mechanisms governing cancer resistance to macrophage phagocytosis.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Figures










References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

