Bioaccumulation of therapeutic drugs by human gut bacteria
- PMID: 34497420
- PMCID: PMC7614428
- DOI: 10.1038/s41586-021-03891-8
Bioaccumulation of therapeutic drugs by human gut bacteria
Abstract
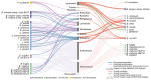
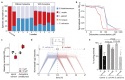
Bacteria in the gut can modulate the availability and efficacy of therapeutic drugs. However, the systematic mapping of the interactions between drugs and bacteria has only started recently1 and the main underlying mechanism proposed is the chemical transformation of drugs by microorganisms (biotransformation). Here we investigated the depletion of 15 structurally diverse drugs by 25 representative strains of gut bacteria. This revealed 70 bacteria-drug interactions, 29 of which had not to our knowledge been reported before. Over half of the new interactions can be ascribed to bioaccumulation; that is, bacteria storing the drug intracellularly without chemically modifying it, and in most cases without the growth of the bacteria being affected. As a case in point, we studied the molecular basis of bioaccumulation of the widely used antidepressant duloxetine by using click chemistry, thermal proteome profiling and metabolomics. We find that duloxetine binds to several metabolic enzymes and changes the metabolite secretion of the respective bacteria. When tested in a defined microbial community of accumulators and non-accumulators, duloxetine markedly altered the composition of the community through metabolic cross-feeding. We further validated our findings in an animal model, showing that bioaccumulating bacteria attenuate the behavioural response of Caenorhabditis elegans to duloxetine. Together, our results show that bioaccumulation by gut bacteria may be a common mechanism that alters drug availability and bacterial metabolism, with implications for microbiota composition, pharmacokinetics, side effects and drug responses, probably in an individual manner.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
M.K., S.A., L.M., M.T., P.B., A.T. and K.R.P. are inventors in a patent application based on the findings reported in this study (US patent application number 16966322). S.B., A.M., P.P., S.D., J.V., B.S., T.S., E.K., D.K., K.Z., E.M., M. Banzhaf, M.M., F.H., L.N., A.R.B., T.B., V.P., M. Kumar, C.S., M. Beck, J.H., M.Z., D.C.S., F.C., and M.M.S. declare no competing interests.
Figures

Comment in
-
Scooping up all the drugs.Nat Rev Microbiol. 2021 Nov;19(11):682. doi: 10.1038/s41579-021-00637-1. Nat Rev Microbiol. 2021. PMID: 34552266 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

