DeepComplex: A Web Server of Predicting Protein Complex Structures by Deep Learning Inter-chain Contact Prediction and Distance-Based Modelling
- PMID: 34497831
- PMCID: PMC8419425
- DOI: 10.3389/fmolb.2021.716973
DeepComplex: A Web Server of Predicting Protein Complex Structures by Deep Learning Inter-chain Contact Prediction and Distance-Based Modelling
Abstract
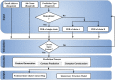
Proteins interact to form complexes. Predicting the quaternary structure of protein complexes is useful for protein function analysis, protein engineering, and drug design. However, few user-friendly tools leveraging the latest deep learning technology for inter-chain contact prediction and the distance-based modelling to predict protein quaternary structures are available. To address this gap, we develop DeepComplex, a web server for predicting structures of dimeric protein complexes. It uses deep learning to predict inter-chain contacts in a homodimer or heterodimer. The predicted contacts are then used to construct a quaternary structure of the dimer by the distance-based modelling, which can be interactively viewed and analysed. The web server is freely accessible and requires no registration. It can be easily used by providing a job name and an email address along with the tertiary structure for one chain of a homodimer or two chains of a heterodimer. The output webpage provides the multiple sequence alignment, predicted inter-chain residue-residue contact map, and predicted quaternary structure of the dimer. DeepComplex web server is freely available at http://tulip.rnet.missouri.edu/deepcomplex/web_index.html.
Keywords: deep learning; distance-based modeling; inter-chain contact prediction; protein complex structure prediction; protein interaction; protein quaternary structure prediction.
Copyright © 2021 Quadir, Roy, Soltanikazemi and Cheng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Distance-based reconstruction of protein quaternary structures from inter-chain contacts.Proteins. 2022 Mar;90(3):720-731. doi: 10.1002/prot.26269. Epub 2021 Nov 2. Proteins. 2022. PMID: 34716620 Free PMC article.
-
Prediction of inter-chain distance maps of protein complexes with 2D attention-based deep neural networks.Nat Commun. 2022 Nov 15;13(1):6963. doi: 10.1038/s41467-022-34600-2. Nat Commun. 2022. PMID: 36379943 Free PMC article.
-
The MULTICOM Protein Structure Prediction Server Empowered by Deep Learning and Contact Distance Prediction.Methods Mol Biol. 2020;2165:13-26. doi: 10.1007/978-1-0716-0708-4_2. Methods Mol Biol. 2020. PMID: 32621217
-
The trRosetta server for fast and accurate protein structure prediction.Nat Protoc. 2021 Dec;16(12):5634-5651. doi: 10.1038/s41596-021-00628-9. Epub 2021 Nov 10. Nat Protoc. 2021. PMID: 34759384 Review.
-
QSalignWeb: A Server to Predict and Analyze Protein Quaternary Structure.Front Mol Biosci. 2022 Jan 5;8:787510. doi: 10.3389/fmolb.2021.787510. eCollection 2021. Front Mol Biosci. 2022. PMID: 35071324 Free PMC article. Review.
Cited by
-
Distance-based reconstruction of protein quaternary structures from inter-chain contacts.Proteins. 2022 Mar;90(3):720-731. doi: 10.1002/prot.26269. Epub 2021 Nov 2. Proteins. 2022. PMID: 34716620 Free PMC article.
-
High-Performance Deep Learning Toolbox for Genome-Scale Prediction of Protein Structure and Function.Workshop Mach Learn HPC Environ. 2021 Nov;2021:46-57. doi: 10.1109/mlhpc54614.2021.00010. Epub 2021 Dec 27. Workshop Mach Learn HPC Environ. 2021. PMID: 35112110 Free PMC article.
-
Prediction of inter-chain distance maps of protein complexes with 2D attention-based deep neural networks.Nat Commun. 2022 Nov 15;13(1):6963. doi: 10.1038/s41467-022-34600-2. Nat Commun. 2022. PMID: 36379943 Free PMC article.
-
Machine Learning Methods in Protein-Protein Docking.Methods Mol Biol. 2024;2780:107-126. doi: 10.1007/978-1-0716-3985-6_7. Methods Mol Biol. 2024. PMID: 38987466
-
Enhancing AlphaFold-Multimer-based Protein Complex Structure Prediction with MULTICOM in CASP15.bioRxiv [Preprint]. 2023 May 18:2023.05.16.541055. doi: 10.1101/2023.05.16.541055. bioRxiv. 2023. Update in: Commun Chem. 2023 Sep 7;6(1):188. doi: 10.1038/s42004-023-00991-6. PMID: 37293073 Free PMC article. Updated. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

