Data-Independent-Acquisition-Based Proteomic Approach towards Understanding the Acclimation Strategy of Oleaginous Microalga Microchloropsis gaditana CCMP526 in Hypersaline Conditions
- PMID: 34497906
- PMCID: PMC8412934
- DOI: 10.1021/acsomega.1c02786
Data-Independent-Acquisition-Based Proteomic Approach towards Understanding the Acclimation Strategy of Oleaginous Microalga Microchloropsis gaditana CCMP526 in Hypersaline Conditions
Abstract
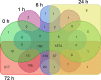
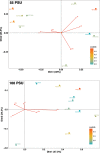
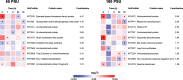
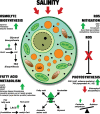
Salinity is one of the significant factors that affect growth and cellular metabolism, including photosynthesis and lipid accumulation, in microalgae and higher plants. Microchloropsis gaditana CCMP526 can acclimatize to different salinity levels by accumulating compatible solutes, carbohydrates, and lipids as energy storage molecules. We used proteomics to understand the molecular basis for acclimation of M. gaditana to increased salinity levels [55 and 100 PSU (practical salinity unit)]. Correspondence analysis was used for the identification of salinity-responsive proteins (SRPs). The highest number of salinity-induced proteins was observed in 100 PSU. Gene ontology enrichment analysis revealed a separate path of acclimation for cells exposed to 55 and 100 PSU. Osmolyte and lipid biosynthesis were upregulated in hypersaline conditions. Concomitantly, lipid oxidation pathways were also upregulated in hypersaline conditions, providing acetyl-CoA for energy metabolism through the tricarboxylic acid cycle. Carbon fixation and photosynthesis were tightly regulated, while chlorophyll biosynthesis was affected in hypersaline conditions. Importantly, temporal proteome analysis of salinity-induced M. gaditana revealed vital SRPs which could be used for engineering salinity resilient microalgal strains for improved productivity in hypersaline culture conditions.
© 2021 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Brennan L.; Owende P. Biofuels from Microalgae—a Review of Technologies for Production, Processing, and Extractions of Biofuels and Co-Products. Renewable Sustainable Energy Rev. 2010, 14, 557–577. 10.1016/j.rser.2009.10.009. - DOI
-
- Doan T. T. Y.; Sivaloganathan B.; Obbard J. P. Screening of Marine Microalgae for Biodiesel Feedstock. Biomass Bioenergy 2011, 35, 2534–2544. 10.1016/j.biombioe.2011.02.021. - DOI
-
- Corteggiani Carpinelli E.; Telatin A.; Vitulo N.; Forcato C.; D’Angelo M.; Schiavon R.; Vezzi A.; Giacometti G. M.; Morosinotto T.; Valle G. Chromosome Scale Genome Assembly and Transcriptome Profiling of Nannochloropsis gaditana in Nitrogen Depletion. Mol. Plant 2014, 7, 323–335. 10.1093/mp/sst120. - DOI - PubMed
-
- Huertas E.; Montero O.; Lubián L. M. Effects of Dissolved Inorganic Carbon Availability on Growth, Nutrient Uptake and Chlorophyll Fluorescence of Two Species of Marine Microalgae. Aquac. Eng. 2000, 22, 181–197. 10.1016/s0144-8609(99)00038-2. - DOI
LinkOut - more resources
Full Text Sources

