High-Throughput Digital Image Analysis Reveals Distinct Patterns of Dystrophin Expression in Dystrophinopathy Patients
- PMID: 34498054
- PMCID: PMC8557329
- DOI: 10.1093/jnen/nlab088
High-Throughput Digital Image Analysis Reveals Distinct Patterns of Dystrophin Expression in Dystrophinopathy Patients
Abstract
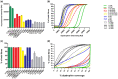
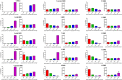
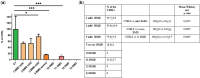
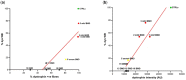
Duchenne muscular dystrophy (DMD) is an incurable disease caused by out-of-frame DMD gene deletions while in frame deletions lead to the milder Becker muscular dystrophy (BMD). In the last decade several antisense oligonucleotides drugs have been developed to induce a partially functional internally deleted dystrophin, similar to that produced in BMD, and expected to ameliorate the disease course. The pattern of dystrophin expression and functionality in dystrophinopathy patients is variable due to multiple factors, such as molecular functionality of the dystrophin and its distribution. To benchmark the success of therapeutic intervention, a clear understanding of dystrophin expression patterns in dystrophinopathy patients is vital. Recently, several groups have used innovative techniques to quantify dystrophin in muscle biopsies of children but not in patients with milder BMD. This study reports on dystrophin expression using both Western blotting and an automated, high-throughput, image analysis platform in DMD, BMD, and intermediate DMD/BMD skeletal muscle biopsies. Our results found a significant correlation between Western blot and immunofluorescent quantification indicating consistency between the different methodologies. However, we identified significant inter- and intradisease heterogeneity of patterns of dystrophin expression in patients irrespective of the amount detected on blot, due to variability in both fluorescence intensity and dystrophin sarcolemmal circumference coverage. Our data highlight the heterogeneity of the pattern of dystrophin expression in BMD, which will assist the assessment of dystrophin restoration therapies.
Keywords: Becker muscular dystrophy; Duchenne muscular dystrophy; Dystrophin; High–throughput digital analysis; Muscle biopsy; Skeletal muscle.
© 2021 American Association of Neuropathologists, Inc.
Figures

References
-
- Flanigan KM. Duchenne and Becker muscular dystrophies. Neurol Clin 2014;32:671–88 - PubMed
-
- Muntoni F, Torelli S, Ferlini A.. Dystrophin and mutations: One gene, several proteins, multiple phenotypes. Lancet Neurol 2003;2:731–40 - PubMed
-
- . Rando TA. The dystrophin-glycoprotein complex, cellular signaling, and the regulation of cell survival in the muscular dystrophies. Muscle Nerve 2001;24:1575–94 - PubMed

