SnRK1 stimulates the histone H3K27me3 demethylase JMJ705 to regulate a transcriptional switch to control energy homeostasis
- PMID: 34498077
- PMCID: PMC8643663
- DOI: 10.1093/plcell/koab224
SnRK1 stimulates the histone H3K27me3 demethylase JMJ705 to regulate a transcriptional switch to control energy homeostasis
Abstract
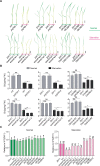
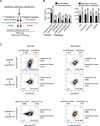
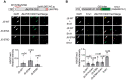
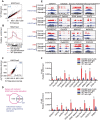
Plant SNF1-Related Kinase1 (SnRK1) is an evolutionarily conserved energy-sensing protein kinase that orchestrates transcriptional networks to maintain cellular energy homeostasis when energy supplies become limited. However, the mechanism by which SnRK1 regulates this gene expression switch to gauge cellular energy status remains largely unclear. In this work, we show that the rice histone H3K27me3 demethylase JMJ705 is required for low energy stress tolerance in rice plants. The genetic inactivation of JMJ705 resulted in similar effects as those of the rice snrk1 mutant on the transcriptome, which impairs not only the promotion of the low energy stress-triggered transcriptional program but also the repression of the program under an energy-sufficient state. We show that the α-subunit of OsSnRK1 interacts with and phosphorylates JMJ705 to stimulate its H3K27me3 demethylase activity. Further analysis revealed that JMJ705 directly targets a set of low energy stress-responsive transcription factor genes. These results uncover the chromatin mechanism of SnRK1-regulated gene expression in both energy-sufficient and -limited states in plants and suggest that JMJ705 functions as an upstream regulator of the SnRK1α-controlled transcriptional network.
© American Society of Plant Biologists 2021. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures







References
-
- Baena-Gonzalez E, Hanson J (2017) Shaping plant development through the SnRK1-TOR metabolic regulators. Curr Opin Plant Biol 35: 152–157 - PubMed
-
- Baena-Gonzalez E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 - PubMed
-
- Broeckx T, Hulsmans S, Rolland F (2016) The plant energy sensor: evolutionary conservation and divergence of SnRK1 structure, regulation, and function. J Exp Bot 67: 6215–6252 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

