Gluconeogenesis in Plants: A Key Interface between Organic Acid/Amino Acid/Lipid and Sugar Metabolism
- PMID: 34500562
- PMCID: PMC8434439
- DOI: 10.3390/molecules26175129
Gluconeogenesis in Plants: A Key Interface between Organic Acid/Amino Acid/Lipid and Sugar Metabolism
Abstract
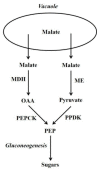
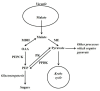
Gluconeogenesis is a key interface between organic acid/amino acid/lipid and sugar metabolism. The aims of this article are four-fold. First, to provide a concise overview of plant gluconeogenesis. Second, to emphasise the widespread occurrence of gluconeogenesis and its utilisation in diverse processes. Third, to stress the importance of the vacuolar storage and release of Krebs cycle acids/nitrogenous compounds, and of the role of gluconeogenesis and malic enzyme in this process. Fourth, to outline the contribution of fine control of enzyme activity to the coordinate-regulation of gluconeogenesis and malate metabolism, and the importance of cytosolic pH in this.
Keywords: gluconeogenesis; malate; malic enzyme; nitrogen metabolism; organic acids; phosphoenolpyruvate carboxykinase; pyruvate orthophosphate dikinase; vacuole.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Famiani F., Farinelli D., Frioni T., Palliotti A., Battistelli A., Moscatello S., Walker R.P. Malate as substrate for catabolism and gluconeogenesis during ripening in the pericarp of different grape cultivars. Biol. Plant. 2016;60:155–162. doi: 10.1007/s10535-015-0574-2. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

