Mechanisms of Binding Specificity among bHLH Transcription Factors
- PMID: 34502060
- PMCID: PMC8431614
- DOI: 10.3390/ijms22179150
Mechanisms of Binding Specificity among bHLH Transcription Factors
Abstract
The transcriptome of every cell is orchestrated by the complex network of interaction between transcription factors (TFs) and their binding sites on DNA. Disruption of this network can result in many forms of organism malfunction but also can be the substrate of positive natural selection. However, understanding the specific determinants of each of these individual TF-DNA interactions is a challenging task as it requires integrating the multiple possible mechanisms by which a given TF ends up interacting with a specific genomic region. These mechanisms include DNA motif preferences, which can be determined by nucleotide sequence but also by DNA's shape; post-translational modifications of the TF, such as phosphorylation; and dimerization partners and co-factors, which can mediate multiple forms of direct or indirect cooperative binding. Binding can also be affected by epigenetic modifications of putative target regions, including DNA methylation and nucleosome occupancy. In this review, we describe how all these mechanisms have a role and crosstalk in one specific family of TFs, the basic helix-loop-helix (bHLH), with a very conserved DNA binding domain and a similar DNA preferred motif, the E-box. Here, we compile and discuss a rich catalog of strategies used by bHLH to acquire TF-specific genome-wide landscapes of binding sites.
Keywords: ChIP-seq; E-box; bHLH; co-factors; dimerization; pioneer factors; transcription factor binding sites.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
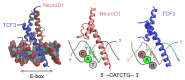


References
-
- Atak Z.K., Taskiran I.I., Demeulemeester J., Flerin C., Mauduit D., Minnoye L., Hulselmans G., Christiaens V., Ghanem G.E., Wouters J., et al. Interpretation of allele-specific chromatin accessibility using cell state–aware deep learning. Genome Res. 2021;31:1082–1096. doi: 10.1101/gr.260851.120. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

