Chemical Chaperones Modulate the Formation of Metabolite Assemblies
- PMID: 34502079
- PMCID: PMC8431448
- DOI: 10.3390/ijms22179172
Chemical Chaperones Modulate the Formation of Metabolite Assemblies
Abstract
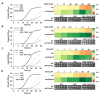
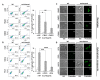
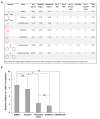
The formation of amyloid-like structures by metabolites is associated with several inborn errors of metabolism (IEMs). These structures display most of the biological, chemical and physical properties of protein amyloids. However, the molecular interactions underlying the assembly remain elusive, and so far, no modulating therapeutic agents are available for clinical use. Chemical chaperones are known to inhibit protein and peptide amyloid formation and stabilize misfolded enzymes. Here, we provide an in-depth characterization of the inhibitory effect of osmolytes and hydrophobic chemical chaperones on metabolite assemblies, thus extending their functional repertoire. We applied a combined in vivo-in vitro-in silico approach and show their ability to inhibit metabolite amyloid-induced toxicity and reduce cellular amyloid content in yeast. We further used various biophysical techniques demonstrating direct inhibition of adenine self-assembly and alteration of fibril morphology by chemical chaperones. Using a scaffold-based approach, we analyzed the physiochemical properties of various dimethyl sulfoxide derivatives and their role in inhibiting metabolite self-assembly. Lastly, we employed whole-atom molecular dynamics simulations to elucidate the role of hydrogen bonds in osmolyte inhibition. Our results imply a dual mode of action of chemical chaperones as IEMs therapeutics, that could be implemented in the rational design of novel lead-like molecules.
Keywords: adenine; amyloid formation; chemical chaperones; hydrophobic compounds; inborn errors of metabolism; metabolite assemblies; osmolytes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Fibril formation and therapeutic targeting of amyloid-like structures in a yeast model of adenine accumulation.Nat Commun. 2019 Jan 8;10(1):62. doi: 10.1038/s41467-018-07966-5. Nat Commun. 2019. PMID: 30622276 Free PMC article.
-
A hydrophobic low-complexity region regulates aggregation of the yeast pyruvate kinase Cdc19 into amyloid-like aggregates in vitro.J Biol Chem. 2018 Jul 20;293(29):11424-11432. doi: 10.1074/jbc.RA117.001628. Epub 2018 May 31. J Biol Chem. 2018. PMID: 29853641 Free PMC article.
-
ATP modulates self-perpetuating conformational conversion generating structurally distinct yeast prion amyloids that limit autocatalytic amplification.J Biol Chem. 2023 May;299(5):104654. doi: 10.1016/j.jbc.2023.104654. Epub 2023 Mar 28. J Biol Chem. 2023. PMID: 36990219 Free PMC article.
-
Impact of Amyloid Polymorphism on Prion-Chaperone Interactions in Yeast.Viruses. 2019 Apr 16;11(4):349. doi: 10.3390/v11040349. Viruses. 2019. PMID: 30995727 Free PMC article. Review.
-
Metabolite assemblies: A surprising extension to the amyloid hypothesis.Curr Opin Chem Biol. 2021 Oct;64:154-164. doi: 10.1016/j.cbpa.2021.07.005. Epub 2021 Sep 2. Curr Opin Chem Biol. 2021. PMID: 34482124 Review.
Cited by
-
Branched-Chain Amino Acid Assembly into Amyloid-like Fibrils Provides a New Paradigm for Maple Syrup Urine Disease Pathology.Int J Mol Sci. 2023 Nov 6;24(21):15999. doi: 10.3390/ijms242115999. Int J Mol Sci. 2023. PMID: 37958982 Free PMC article.
-
The rise and fall of adenine clusters in the gas phase: a glimpse into crystal growth and nucleation.Anal Bioanal Chem. 2024 Sep;416(23):5037-5048. doi: 10.1007/s00216-024-05442-2. Epub 2024 Jul 20. Anal Bioanal Chem. 2024. PMID: 39031229
-
Functional Activities of Cohesin Proteins Can Be Altered by Chemical Chaperones.Protein J. 2025 Jul 16. doi: 10.1007/s10930-025-10276-7. Online ahead of print. Protein J. 2025. PMID: 40668332
-
USP10 as a Potential Therapeutic Target in Human Cancers.Genes (Basel). 2022 May 6;13(5):831. doi: 10.3390/genes13050831. Genes (Basel). 2022. PMID: 35627217 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

