Eugenol-Induced Autophagy and Apoptosis in Breast Cancer Cells via PI3K/AKT/FOXO3a Pathway Inhibition
- PMID: 34502165
- PMCID: PMC8430664
- DOI: 10.3390/ijms22179243
Eugenol-Induced Autophagy and Apoptosis in Breast Cancer Cells via PI3K/AKT/FOXO3a Pathway Inhibition
Abstract
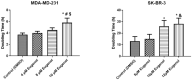
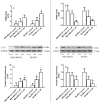
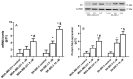
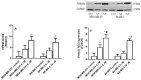
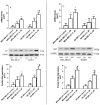
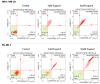
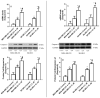
The use of natural compounds is promising in approaches to prevent and treat cancer. The long-term application of most currently employed chemotherapy techniques has toxic side effects. Eugenol, a phenolic phytochemical extracted from certain essential oils, has an anti-cancer effect. The modulation of autophagy can promote either the survival or apoptosis of cancer cells. Triple-negative (MDA-MB-231) and HER2 positive (SK-BR-3) breast cancer cell lines were treated with different doses of eugenol. Apoptosis was detected by a flow-cytometry technique, while autophagy was detected by acridine orange. Real-time PCR and Western blot assays were applied to investigate the effect of eugenol on the gene and protein expression levels of autophagy and apoptotic genes. Treating cells with different concentrations of eugenol significantly inhibited cell proliferation. The protein levels of AKT serine/threonine kinase 1 (AKT), forkhead box O3 (FOXO3a), cyclin dependent kinase inhibitor 1A (p21), cyclin-dependent kinase inhibitor (p27), and Caspase-3 and -9 increased significantly in Eugenol-treated cells. Eugenol also induced autophagy by upregulating the expression levels of microtubule-associated protein 1 light chain 3 (LC3) and downregulating the expression of nucleoporin 62 (NU p62). Eugenol is a promising natural anti-cancer agent against triple-negative and HER2-positive breast cancer. It appears to work by targeting the caspase pathway and by inducing autophagic cell death.
Keywords: PI3K/AKT/FOXO3a pathway; autophagy; eugenol; triple negative breast cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
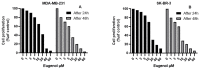

References
-
- Maayah Z.H., Ghebeh H., Alhaider A.A., El-Kadi A.O., Soshilov A.A., Denison M.S., Ansari M.A., Korashy H.M. Metformin inhibits 7,12-dimethylbenz[a]anthracene-induced breast carcinogenesis and adduct formation in human breast cells by inhibiting the cytochrome P4501A1/aryl hydrocarbon receptor signaling pathway. Toxicol. Appl. Pharmacol. 2015;284:217–226. doi: 10.1016/j.taap.2015.02.007. - DOI - PubMed
-
- Rychlý B., Sidlová H., Daniś D. The 2007 World Health Organisation classification of tumours of the central nervous system, comparison with 2000 classification. Ceskoslovenska Patol. 2008;44:35–36. - PubMed
-
- Korlimarla A., Prabhu J.S., Remacle J., Rajarajan S., Raja U., Anupama C.E., Srinath B.S., Manjunath S., Gopinath K.S., Correa M., et al. Identification of BRCA1 Deficiency Using Multi-Analyte Estimation of BRCA1 and Its Repressors in FFPE Tumor Samples from Patients with Triple Negative Breast Cancer. PLoS ONE. 2016;11:e0153113. doi: 10.1371/journal.pone.0153113. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

