Ptk7 Is Dynamically Localized at Neural Crest Cell-Cell Contact Sites and Functions in Contact Inhibition of Locomotion
- PMID: 34502237
- PMCID: PMC8431534
- DOI: 10.3390/ijms22179324
Ptk7 Is Dynamically Localized at Neural Crest Cell-Cell Contact Sites and Functions in Contact Inhibition of Locomotion
Abstract
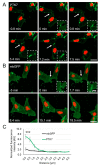
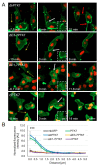
Neural crest (NC) cells are highly migratory cells that contribute to various vertebrate tissues, and whose migratory behaviors resemble cancer cell migration and invasion. Information exchange via dynamic NC cell-cell contact is one mechanism by which the directionality of migrating NC cells is controlled. One transmembrane protein that is most likely involved in this process is protein tyrosine kinase 7 (PTK7), an evolutionary conserved Wnt co-receptor that is expressed in cranial NC cells and several tumor cells. In Xenopus, Ptk7 is required for NC migration. In this study, we show that the Ptk7 protein is dynamically localized at cell-cell contact zones of migrating Xenopus NC cells and required for contact inhibition of locomotion (CIL). Using deletion constructs of Ptk7, we determined that the extracellular immunoglobulin domains of Ptk7 are important for its transient accumulation and that they mediate homophilic binding. Conversely, we found that ectopic expression of Ptk7 in non-NC cells was able to prevent NC cell invasion. However, deletion of the extracellular domains of Ptk7 abolished this effect. Thus, Ptk7 is sufficient at protecting non-NC tissue from NC cell invasion, suggesting a common role of PTK7 in contact inhibition, cell invasion, and tissue integrity.
Keywords: PTK7; cell invasion; dynamic cell–cell contacts; neural crest migration.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

