ZFP36L2 Role in Thyroid Functionality
- PMID: 34502288
- PMCID: PMC8431063
- DOI: 10.3390/ijms22179379
ZFP36L2 Role in Thyroid Functionality
Abstract
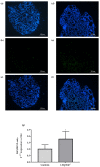
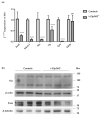
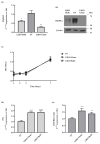
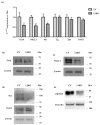
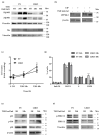
Thyroid hormone levels are usually genetically determined. Thyrocytes produce a unique set of enzymes that are dedicated to thyroid hormone synthesis. While thyroid transcriptional regulation is well-characterized, post-transcriptional mechanisms have been less investigated. Here, we describe the involvement of ZFP36L2, a protein that stimulates degradation of target mRNAs, in thyroid development and function, by in vivo and in vitro gene targeting in thyrocytes. Thyroid-specific Zfp36l2-/- females were hypothyroid, with reduced levels of circulating free Thyroxine (cfT4) and Triiodothyronine (cfT3). Their hypothyroidism was due to dyshormonogenesis, already evident one week after weaning, while thyroid development appeared normal. We observed decreases in several thyroid-specific transcripts and proteins, such as Nis and its transcriptional regulators (Pax8 and Nkx2.1), and increased apoptosis in Zfp36l2-/- thyroids. Nis, Pax8, and Nkx2.1 mRNAs were also reduced in Zfp36l2 knock-out thyrocytes in vitro (L2KO), in which we confirmed the increased apoptosis. Finally, in L2KO cells, we showed an altered response to TSH stimulation regarding both thyroid-specific gene expression and cell proliferation and survival. This result was supported by increases in P21/WAF1 and p-P38MAPK levels. Mechanistically, we confirmed Notch1 as a target of ZFP36L2 in the thyroid since its levels were increased in both in vitro and in vivo models. In both models, the levels of Id4 mRNA, a potential inhibitor of Pax8 activity, were increased. Overall, the data indicate that the regulation of mRNA stability by ZFP36L2 is a mechanism that controls the function and survival of thyrocytes.
Keywords: Nis; Notch1; RNA stability; Zfp36l2 KO; apoptosis; hypothyroidism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Grants and funding
- 12010/Istituto Nazionale per l'Assicurazione Contro Gli Infortuni sul Lavoro
- 23/Sensor Regione Campania
- POR Campania FESR 2014/2020 O.S. 1.1 Az. 1.1.3 E 1.1.4-CUP 572 B63D18000150007/Goodwater Regione Campania
- RARE PLATNET, SATIN and COEPICA/POR FESR 2014-2020 Regione Campania
- Intramural Research Program/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

