Comparative Proteomic Analysis of Tolerant and Sensitive Varieties Reveals That Phenylpropanoid Biosynthesis Contributes to Salt Tolerance in Mulberry
- PMID: 34502318
- PMCID: PMC8431035
- DOI: 10.3390/ijms22179402
Comparative Proteomic Analysis of Tolerant and Sensitive Varieties Reveals That Phenylpropanoid Biosynthesis Contributes to Salt Tolerance in Mulberry
Abstract
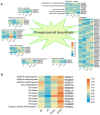
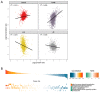
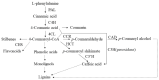
Mulberry, an important woody tree, has strong tolerance to environmental stresses, including salinity, drought, and heavy metal stress. However, the current research on mulberry resistance focuses mainly on the selection of resistant resources and the determination of physiological indicators. In order to clarify the molecular mechanism of salt tolerance in mulberry, the physiological changes and proteomic profiles were comprehensively analyzed in salt-tolerant (Jisang3) and salt-sensitive (Guisangyou12) mulberry varieties. After salt treatment, the malondialdehyde (MDA) content and proline content were significantly increased compared to control, and the MDA and proline content in G12 was significantly lower than in Jisang3 under salt stress. The calcium content was significantly reduced in the salt-sensitive mulberry varieties Guisangyou12 (G12), while sodium content was significantly increased in both mulberry varieties. Although the Jisang3 is salt-tolerant, salt stress caused more reductions of photosynthetic rate in Jisang3 than Guisangyou12. Using tandem mass tags (TMT)-based proteomics, the changes of mulberry proteome levels were analyzed in salt-tolerant and salt-sensitive mulberry varieties under salt stress. Combined with GO and KEGG databases, the differentially expressed proteins were significantly enriched in the GO terms of amino acid transport and metabolism and posttranslational modification, protein turnover up-classified in Guisangyou12 while down-classified in Jisang3. Through the comparison of proteomic level, we identified the phenylpropanoid biosynthesis may play an important role in salt tolerance of mulberry. We clarified the molecular mechanism of mulberry salt tolerance, which is of great significance for the selection of excellent candidate genes for saline-alkali soil management and mulberry stress resistance genetic engineering.
Keywords: TMT proteomics; mulberry; phenylpropanoid metabolism; salt stress.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

