Genome-Wide Identification and Characterization of Short-Chain Dehydrogenase/Reductase (SDR) Gene Family in Medicago truncatula
- PMID: 34502406
- PMCID: PMC8430790
- DOI: 10.3390/ijms22179498
Genome-Wide Identification and Characterization of Short-Chain Dehydrogenase/Reductase (SDR) Gene Family in Medicago truncatula
Abstract
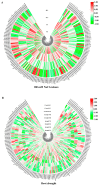
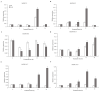
Short-chain dehydrogenase/reductase (SDR) belongs to the NAD(P)(H)-dependent oxidoreductase superfamily. Limited investigations reveal that SDRs participate in diverse metabolisms. A genome-wide identification of the SDR gene family in M. truncatula was conducted. A total of 213 MtSDR genes were identified, and they were distributed on all chromosomes unevenly. MtSDR proteins were categorized into seven subgroups based on phylogenetic analysis and three types including 'classic', 'extended', and 'atypical', depending on the cofactor-binding site and active site. Analysis of the data from M. truncatula Gene Expression Atlas (MtGEA) showed that above half of MtSDRs were expressed in at least one organ, and lots of MtSDRs had a preference in a tissue-specific expression. The cis-acting element responsive to plant hormones (salicylic acid, ABA, auxin, MeJA, and gibberellin) and stresses were found in the promoter of some MtSDRs. Many genes of MtSDR7C,MtSDR65C, MtSDR110C, MtSDR114C, and MtSDR108E families were responsive to drought, salt, and cold. The study provides useful information for further investigation on biological functions of MtSDRs, especially in abiotic stress adaptation, in the future.
Keywords: Medicago truncatula; abiotic stress; cis-acting elements; expression profiles; short-chain dehydrogenase/reductase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Genome-Wide Analysis and Characterization of the SDR Gene Superfamily in Cinnamomum camphora and Identification of Synthase for Eugenol Biosynthesis.Int J Mol Sci. 2024 Sep 19;25(18):10084. doi: 10.3390/ijms251810084. Int J Mol Sci. 2024. PMID: 39337570 Free PMC article.
-
Calmodulin-Like (CML) Gene Family in Medicago truncatula: Genome-Wide Identification, Characterization and Expression Analysis.Int J Mol Sci. 2020 Sep 27;21(19):7142. doi: 10.3390/ijms21197142. Int J Mol Sci. 2020. PMID: 32992668 Free PMC article.
-
Genome-wide analysis of the KCS gene family in Medicago truncatula and their expression profile under various abiotic stress.Sci Rep. 2025 May 7;15(1):15938. doi: 10.1038/s41598-025-00809-6. Sci Rep. 2025. PMID: 40335581 Free PMC article.
-
A genome-wide study of the lipoxygenase gene families in Medicago truncatula and Medicago sativa reveals that MtLOX24 participates in the methyl jasmonate response.BMC Genomics. 2024 Feb 19;25(1):195. doi: 10.1186/s12864-024-10071-1. BMC Genomics. 2024. PMID: 38373903 Free PMC article.
-
Genome-Wide Identification and Expression Profiling Analysis of the Trihelix Gene Family Under Abiotic Stresses in Medicago truncatula.Genes (Basel). 2020 Nov 23;11(11):1389. doi: 10.3390/genes11111389. Genes (Basel). 2020. PMID: 33238556 Free PMC article.
Cited by
-
Silicon-Induced Tolerance against Arsenic Toxicity by Activating Physiological, Anatomical and Biochemical Regulation in Phoenix dactylifera (Date Palm).Plants (Basel). 2022 Aug 31;11(17):2263. doi: 10.3390/plants11172263. Plants (Basel). 2022. PMID: 36079645 Free PMC article.
-
Uncovering the transcriptional responses of tobacco (Nicotiana tabacum L.) roots to Ralstonia solanacearum infection: a comparative study of resistant and susceptible cultivars.BMC Plant Biol. 2023 Dec 6;23(1):620. doi: 10.1186/s12870-023-04633-w. BMC Plant Biol. 2023. PMID: 38057713 Free PMC article.
-
Isolation of the 3β-HSD promoter from Digitalis ferruginea subsp. ferruginea and its functional characterization in Arabidopsis thaliana.Mol Biol Rep. 2022 Jul;49(7):7173-7183. doi: 10.1007/s11033-022-07634-4. Epub 2022 Jun 22. Mol Biol Rep. 2022. PMID: 35733064
-
Genome-Wide Analysis and Characterization of the SDR Gene Superfamily in Cinnamomum camphora and Identification of Synthase for Eugenol Biosynthesis.Int J Mol Sci. 2024 Sep 19;25(18):10084. doi: 10.3390/ijms251810084. Int J Mol Sci. 2024. PMID: 39337570 Free PMC article.
-
Genome-Wide Analysis of the MADS-Box Gene Family in Hibiscus syriacus and Their Role in Floral Organ Development.Int J Mol Sci. 2023 Dec 28;25(1):406. doi: 10.3390/ijms25010406. Int J Mol Sci. 2023. PMID: 38203576 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

