Docking to a Basic Helix Promotes Specific Phosphorylation by G1-Cdk1
- PMID: 34502421
- PMCID: PMC8431026
- DOI: 10.3390/ijms22179514
Docking to a Basic Helix Promotes Specific Phosphorylation by G1-Cdk1
Abstract
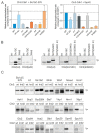
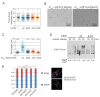
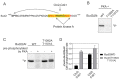
Cyclins are the activators of cyclin-dependent kinase (CDK) complex, but they also act as docking scaffolds for different short linear motifs (SLiMs) in CDK substrates and inhibitors. According to the unified model of CDK function, the cell cycle is coordinated by CDK both via general CDK activity thresholds and cyclin-specific substrate docking. Recently, it was found that the G1-cyclins of S. cerevisiae have a specific function in promoting polarization and growth of the buds, making the G1 cyclins essential for cell survival. Thus, while a uniform CDK specificity of a single cyclin can be sufficient to drive the cell cycle in some cells, such as in fission yeast, cyclin specificity can be essential in other organisms. However, the known G1-CDK specific LP docking motif, was not responsible for this essential function, indicating that G1-CDKs use yet other unknown docking mechanisms. Here we report a discovery of a G1 cyclin-specific (Cln1,2) lysine-arginine-rich helical docking motif (the K/R motif) in G1-CDK targets involved in the mating pathway (Ste7), transcription (Xbp1), bud morphogenesis (Bud2) and spindle pole body (Spc29, Spc42, Spc110, Sli15) function of S. cerevisiae. We also show that the docking efficiency of K/R motif can be regulated by basophilic kinases such as protein kinase A. Our results further widen the list of cyclin specificity mechanisms and may explain the recently demonstrated unique essential function of G1 cyclins in budding yeast.
Keywords: SLiM; cyclin specificity; cyclin-dependent kinase; kinase specificity; phosphorylation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Cyclin-specific docking motifs promote phosphorylation of yeast signaling proteins by G1/S Cdk complexes.Curr Biol. 2011 Oct 11;21(19):1615-23. doi: 10.1016/j.cub.2011.08.033. Epub 2011 Sep 22. Curr Biol. 2011. PMID: 21945277 Free PMC article.
-
Comprehensive Analysis of G1 Cyclin Docking Motif Sequences that Control CDK Regulatory Potency In Vivo.Curr Biol. 2020 Nov 16;30(22):4454-4466.e5. doi: 10.1016/j.cub.2020.08.099. Epub 2020 Sep 24. Curr Biol. 2020. PMID: 32976810 Free PMC article.
-
Cyclin-Specific Docking Mechanisms Reveal the Complexity of M-CDK Function in the Cell Cycle.Mol Cell. 2019 Jul 11;75(1):76-89.e3. doi: 10.1016/j.molcel.2019.04.026. Epub 2019 May 14. Mol Cell. 2019. PMID: 31101497 Free PMC article.
-
CDK signaling via nonconventional CDK phosphorylation sites.Mol Biol Cell. 2023 Nov 1;34(12):pe5. doi: 10.1091/mbc.E22-06-0196. Mol Biol Cell. 2023. PMID: 37906435 Free PMC article. Review.
-
[Molecular mechanisms controlling the cell cycle: fundamental aspects and implications for oncology].Cancer Radiother. 2001 Apr;5(2):109-29. doi: 10.1016/s1278-3218(01)00087-7. Cancer Radiother. 2001. PMID: 11355576 Review. French.
Cited by
-
Integrated analysis of SR-like protein kinases Sky1 and Sky2 links signaling networks with transcriptional regulation in Candida albicans.Front Cell Infect Microbiol. 2023 Apr 3;13:1108235. doi: 10.3389/fcimb.2023.1108235. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37082713 Free PMC article.
-
Emerging approaches to CDK inhibitor development, a structural perspective.RSC Chem Biol. 2022 Dec 14;4(2):146-164. doi: 10.1039/d2cb00201a. eCollection 2023 Feb 8. RSC Chem Biol. 2022. PMID: 36794018 Free PMC article.
References
-
- Morgan D.O. The Cell Cycle: Principles of Control. New Science Press; London, UK: 2007.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

