Relationship between the Phenylpropanoid Pathway and Dwarfism of Paspalum seashore Based on RNA-Seq and iTRAQ
- PMID: 34502485
- PMCID: PMC8431245
- DOI: 10.3390/ijms22179568
Relationship between the Phenylpropanoid Pathway and Dwarfism of Paspalum seashore Based on RNA-Seq and iTRAQ
Abstract
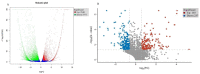
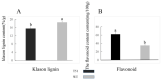
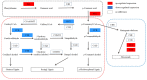
Seashore paspalum is a major warm-season turfgrass requiring frequent mowing. The use of dwarf cultivars with slow growth is a promising method to decrease mowing frequency. The present study was conducted to provide an in-depth understanding of the molecular mechanism of T51 dwarfing in the phenylpropane pathway and to screen the key genes related to dwarfing. For this purpose, we obtained transcriptomic information based on RNA-Seq and proteomic information based on iTRAQ for the dwarf mutant T51 of seashore paspalum. The combined results of transcriptomic and proteomic analysis were used to identify the differential expression pattern of genes at the translational and transcriptional levels. A total of 8311 DEGs were detected at the transcription level, of which 2540 were upregulated and 5771 were downregulated. Based on the transcripts, 2910 proteins were identified using iTRAQ, of which 392 (155 upregulated and 237 downregulated) were DEPs. The phenylpropane pathway was found to be significantly enriched at both the transcriptional and translational levels. Combined with the decrease in lignin content and the increase in flavonoid content in T51, we found that the dwarf phenotype of T51 is closely related to the abnormal synthesis of lignin and flavonoids in the phenylpropane pathway. CCR and HCT may be the key genes for T51 dwarf. This study provides the basis for further study on the dwarfing mechanism of seashore paspalum. The screening of key genes lays a foundation for further studies on the molecular mechanism of seashore paspalum dwarfing.
Keywords: Paspalum seashore; RNA-Seq; dwarfism; iTRAQ; lignin; phenylpropanoid.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Comparative transcriptome profiling provides insights into plant salt tolerance in seashore paspalum (Paspalum vaginatum).BMC Genomics. 2020 Feb 7;21(1):131. doi: 10.1186/s12864-020-6508-1. BMC Genomics. 2020. PMID: 32033524 Free PMC article.
-
RNA-Seq Transcriptomics and iTRAQ Proteomics Analysis Reveal the Dwarfing Mechanism of Blue Fescue (Festuca glauca).Plants (Basel). 2024 Nov 29;13(23):3357. doi: 10.3390/plants13233357. Plants (Basel). 2024. PMID: 39683150 Free PMC article.
-
RNA-Seq and iTRAQ Reveal the Dwarfing Mechanism of Dwarf Polish Wheat (Triticum polonicum L.).Int J Biol Sci. 2016 Apr 8;12(6):653-66. doi: 10.7150/ijbs.14577. eCollection 2016. Int J Biol Sci. 2016. PMID: 27194943 Free PMC article.
-
Genome-Wide Identification and Expression Analysis of NAC Gene Family Members in Seashore Paspalum Under Salt Stress.Plants (Basel). 2024 Dec 23;13(24):3595. doi: 10.3390/plants13243595. Plants (Basel). 2024. PMID: 39771292 Free PMC article.
-
Identification and Validation of Reference Genes for Seashore Paspalum Response to Abiotic Stresses.Int J Mol Sci. 2017 Jun 21;18(6):1322. doi: 10.3390/ijms18061322. Int J Mol Sci. 2017. PMID: 28635628 Free PMC article.
Cited by
-
Total Flavonoid Contents and the Expression of Flavonoid Biosynthetic Genes in Breadfruit (Artocarpus altilis) Scions Growing on Lakoocha (Artocarpus lakoocha) Rootstocks.Plants (Basel). 2023 Sep 16;12(18):3285. doi: 10.3390/plants12183285. Plants (Basel). 2023. PMID: 37765449 Free PMC article.
-
Integrated transcriptomic and metabolomic analysis of flavonoid biosynthesis in cigar tobacco leaves under variable nitrogen regimes.Front Plant Sci. 2025 Jun 23;16:1589215. doi: 10.3389/fpls.2025.1589215. eCollection 2025. Front Plant Sci. 2025. PMID: 40630733 Free PMC article.
-
Integrated metabolomic and transcriptomic analyses of regulatory mechanisms associated with uniconazole-induced dwarfism in banana.BMC Plant Biol. 2022 Dec 28;22(1):614. doi: 10.1186/s12870-022-04005-w. BMC Plant Biol. 2022. PMID: 36575388 Free PMC article.
-
Transcriptome Analysis Reveals the Stress Tolerance to and Accumulation Mechanisms of Cadmium in Paspalum vaginatum Swartz.Plants (Basel). 2022 Aug 9;11(16):2078. doi: 10.3390/plants11162078. Plants (Basel). 2022. PMID: 36015382 Free PMC article.
-
Targeted Metabolic and Transcriptomic Analysis of Pinus yunnanensis var. pygmaea with Loss of Apical Dominance.Curr Issues Mol Biol. 2022 Nov 3;44(11):5485-5497. doi: 10.3390/cimb44110371. Curr Issues Mol Biol. 2022. PMID: 36354683 Free PMC article.
References
-
- Cyril J., Powell G., Duncan R., Baird W. Changes in membrane polar lipid fatty acids of seashore paspalum in response to low temperature exposure. Crop Sci. 2002;42:2031–2037. doi: 10.2135/cropsci2002.2031. - DOI
-
- Lee G., Carrow R.N., Duncan R.R. Salinity tolerance of selected seashore paspalums and bermudagrasses: Root and verdure responses and criteria. HortScience. 2004;39:1143–1147. doi: 10.21273/HORTSCI.39.5.1143. - DOI
-
- Zwar J.A., Chandler P.M. α-Amylase production and leaf protein synthesis in a gibberellin-responsive dwarf mutant of ‘Himalaya’barley (Hordeum vulgare L.) Planta. 1995;197:39–48. doi: 10.1007/BF00239937. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources

