Best Practices for Spatial Profiling for Breast Cancer Research with the GeoMx® Digital Spatial Profiler
- PMID: 34503266
- PMCID: PMC8431590
- DOI: 10.3390/cancers13174456
Best Practices for Spatial Profiling for Breast Cancer Research with the GeoMx® Digital Spatial Profiler
Abstract
Breast cancer is a heterogenous disease with variability in tumor cells and in the surrounding tumor microenvironment (TME). Understanding the molecular diversity in breast cancer is critical for improving prediction of therapeutic response and prognostication. High-plex spatial profiling of tumors enables characterization of heterogeneity in the breast TME, which can holistically illuminate the biology of tumor growth, dissemination and, ultimately, response to therapy. The GeoMx Digital Spatial Profiler (DSP) enables researchers to spatially resolve and quantify proteins and RNA transcripts from tissue sections. The platform is compatible with both formalin-fixed paraffin-embedded and frozen tissues. RNA profiling was developed at the whole transcriptome level for human and mouse samples and protein profiling of 100-plex for human samples. Tissue can be optically segmented for analysis of regions of interest or cell populations to study biology-directed tissue characterization. The GeoMx Breast Cancer Consortium (GBCC) is composed of breast cancer researchers who are developing innovative approaches for spatial profiling to accelerate biomarker discovery. Here, the GBCC presents best practices for GeoMx profiling to promote the collection of high-quality data, optimization of data analysis and integration of datasets to advance collaboration and meta-analyses. Although the capabilities of the platform are presented in the context of breast cancer research, they can be generalized to a variety of other tumor types that are characterized by high heterogeneity.
Keywords: GeoMx; RNA and protein profiling; biomarker discovery; breast cancer; cancer transcriptome atlas; digital spatial profiler; spatial biology; tumor heterogeneity; tumor microenvironment; whole transcriptome atlas.
Conflict of interest statement
A.C. is an employee of ESSA Pharma Inc., a consultant for NanoString Technologies Inc., Refuge Biotechnologies Inc., Bayer AG and Kiromic BioPharma Inc. and is on the scientific advisory board for Arch Oncology, Refuge Biotechnologies Inc. and IO Biotech ApS. M.C.U.C. has an advisory role of Veracyte and is co-inventor of the PAM50 bioclassifier patent with royalties paid by and research funding from NanoString Technologies Inc. S.E.C., P.D., C.A.F., J.G., M.L.H., Y.L., J.P., J.W.R., H.S., S.E.W. are employees of NanoString Technologies Inc., hold NanoString stock or stock options. S.G. receives research funding from Eli Lilly and G1 Therapeutics and honoraria for consulting from Eli Lilly, G1 Therapeutics, Pfizer and Novartis. J.L.G. is a consultant for GlaxoSmithKline (GSK), Codagenix, Verseau, Kymera and Array BioPharma and receives sponsored research support from GSK, Array BioPharma and Eli Lilly. E.A.M. is SAB for Exact Sciences, Merck and Roche/Genentech (compensated) and part of steering committee for Roche/Genentech and Bristol-Myers Squibb (uncompensated). A.P. has an advisory role and research funding from NanoString Technologies Inc. L.P. has received consulting fees and honoraria from Seagen, Pfizer, Astra Zeneca, Merck, Novartis, Bristol-Myers Squibb, Pfizer, Genentech, Eisai, Pieris, Immunomedics, Clovis, Syndax, H3Bio, Radius Health, Daiichi and institutional research funding from Seagen, AstraZeneca, Merck, Pfizer and Bristol Myers Squibb. O.S. is a co-founder of OncoCube Therapeutics LLC, founder and president of LoxiGen, Inc. and a grant recipient from Halozyme Therapeutics Inc. S.M.S. is consulting/honoraria for AstraZeneca, Daiichi Sankyo, Genentech/Hoffmann-La Roche, Lilly Pharmaceuticals, Exact Sciences, Athenex, Bejing Medical Award Foundation, third party writing (Genentech/Hoffman-la Roche), Independent Data Monitoring committee for AstraZeneca and research to institution Kailos Genetics. A.S. has received research support, consulting fees and conference travel support from NanoString Technologies Inc. S.M.T. receives institutional research funding from AstraZeneca, Lilly, Merck, Nektar, Novartis, Pfizer, Genentech/Roche, Immunomedics, Exelixis, Bristol-Myers Squibb, Eisai, Nanostring, Cyclacel, Odonate and Seattle Genetics and has served as an advisor/consultant to AstraZeneca, Lilly, Merck, Nektar, Novartis, Pfizer, Genentech/Roche, Immunomedics, Bristol-Myers Squibb, Eisai, Nanostring, Puma, Sanofi, Celldex, Paxman, Puma, Silverback Therapeutics, G1 Therapeutics, Gilead, AbbVie, Anthenex, OncoPep, Outcomes4Me, Kyowa Kirin Pharmaceuticals, Daiichi-Sankyo, Ellipsis and Samsung Bioepsis Inc. H.B., J.M.C., E.S.H., H.K., J.L., Y.R., J.K.R., I.S., T.S., D.G.S., E..AT.: No competing interests declared.
Figures

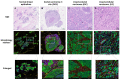

References
-
- Prat A., Cheang M.C., Galván P., Nuciforo P., Paré L., Adamo B., Muñoz M., Viladot M., Press M.F., Gagnon R., et al. Prognostic Value of Intrinsic Subtypes in Hormone Re-ceptor-Positive Metastatic Breast Cancer Treated with Letrozole with or without Lapatinib. JAMA Oncol. 2016;2:1287–1294. doi: 10.1001/jamaoncol.2016.0922. - DOI - PubMed
-
- Prat A., Cheang M.C., Martín M., Parker J.S., Carrasco E., Caballero R., Tyldesley S., Gelmon K., Bernard P.S., Nielsen T.O., et al. Prognostic significance of progesterone recep-tor-positive tumor cells within immunohistochemically defined luminal A breast cancer. J. Clin. Oncol. 2013;31:203–209. doi: 10.1200/JCO.2012.43.4134. - DOI - PMC - PubMed
-
- Akbar M.W., Isbilen M., Belder N., Canlı S.D., Kucukkaraduman B., Turk C., Özgür S., Gure A.O. A Stemness and EMT Based Gene Expression Signature Identifies Phenotypic Plasticity and is A Predictive but Not Prognostic Biomarker for Breast Cancer. J. Cancer. 2020;11:949–961. doi: 10.7150/jca.34649. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

